
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 24,321,123 – 24,321,238 |
| Length | 115 |
| Max. P | 0.967076 |

| Location | 24,321,123 – 24,321,238 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 73.58 |
| Mean single sequence MFE | -33.39 |
| Consensus MFE | -15.51 |
| Energy contribution | -16.74 |
| Covariance contribution | 1.23 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.46 |
| SVM decision value | 1.60 |
| SVM RNA-class probability | 0.967076 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
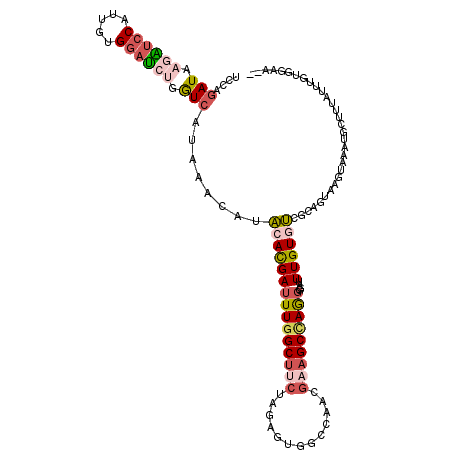
>3R_DroMel_CAF1 24321123 115 + 27905053 UCCAGAUAAGAUCCAUUGUGGAUCUGGUCAUAAACAUACACGAUUUGGCUUCUAGAGUGGCGAACGAAGCCAGGGGAUUUGUGUCGCAGUAAGUAAUUGCUUUGUUUGUGGAAUG (((.(((.((((((.....)))))).)))(((((((.(((((((((((((((.............)))))))))....)))))).(((((.....)))))..))))))))))... ( -37.52) >DroVir_CAF1 123112 100 + 1 UGAGCAUCAAGGACAUAGUGAAUCUGGUCAUAAACGUACACGAAUUGGCCGCAAAAGUCGUGGCCGAAGCAAAAGGCUUUGUGUCGCAGUGAGUACAA-AU-------------- ((.((.(((..(((.(((.....)))))).....((.(((((((((((((((.......)))))))).((.....)))))))))))...))))).)).-..-------------- ( -27.10) >DroPse_CAF1 151795 98 + 1 UCCAGAUGAAUUCCGUAGUGGAGCUCAUCAUAAAUAUUCACGAUUUGGCUGGGAGAAUUGCCAUCGAAGCCAAGGAAUUUGUCACGCAGUAAGUGAC---------------A-- (((.(((((.((((.....)))).)))))...............((((((..((.........))..)))))))))...((((((.......)))))---------------)-- ( -29.00) >DroSec_CAF1 143450 112 + 1 UUCAGAUAAGAUCCAUUGUGGAUCUGGUCAUAAACAUACACGAUUUGGCUUCUAGAGUGGCGAACGAAGCCAGGGCGUUUGUGUCGCAGUGAGUAAUUGCUUUAUUUGUGGA--- ....(((.((((((.....)))))).)))....(((((.(((.(((((((((.............))))))))).))).)))))(((((.((((....))))...)))))..--- ( -35.92) >DroEre_CAF1 152740 113 + 1 UCCAGAUACGAUCCAUUGUGGAUCUGGUCAUAAACAUACAUGAUUUGGCUUCUAGAGUGGCCAACGAAGCCAGGGGAUUUGUAUCCCAGUAAGUAGAAGCCUUAUUUGUGGAA-- (((.(((..(((((.....)))))..)))........(((.(((..((((((((...((((.......)))).(((((....)))))......))))))))..))))))))).-- ( -38.60) >DroYak_CAF1 148934 113 + 1 UCCAGAUACGAUCCAUUGUGGAUCUGGUCAUAAACAUACACGAUUUGGCUUCUAGAGUGGCCAACGAAGCUAGGGGAUUUGUGUCCCAGUGAGUAAAUGAUUUAUUUGUGAAA-- ((((((((.(((((.....))))).((((((..((.(((.((..((((((........)))))))).......(((((....))))).))).))..)))))))))))).))..-- ( -32.20) >consensus UCCAGAUAAGAUCCAUUGUGGAUCUGGUCAUAAACAUACACGAUUUGGCUUCUAGAGUGGCCAACGAAGCCAGGGGAUUUGUGUCGCAGUAAGUAAAUGCUUUAUUUGUGGAA__ ....(((.((((((.....)))))).)))........(((((((((((((((.............)))))))))....))))))............................... (-15.51 = -16.74 + 1.23)


| Location | 24,321,123 – 24,321,238 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 73.58 |
| Mean single sequence MFE | -26.77 |
| Consensus MFE | -12.03 |
| Energy contribution | -13.04 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.45 |
| SVM decision value | 1.28 |
| SVM RNA-class probability | 0.939397 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24321123 115 - 27905053 CAUUCCACAAACAAAGCAAUUACUUACUGCGACACAAAUCCCCUGGCUUCGUUCGCCACUCUAGAAGCCAAAUCGUGUAUGUUUAUGACCAGAUCCACAAUGGAUCUUAUCUGGA ...((((.(((((..(((.........))).((((........(((((((.............)))))))....)))).)))))..((..((((((.....))))))..)))))) ( -27.02) >DroVir_CAF1 123112 100 - 1 --------------AU-UUGUACUCACUGCGACACAAAGCCUUUUGCUUCGGCCACGACUUUUGCGGCCAAUUCGUGUACGUUUAUGACCAGAUUCACUAUGUCCUUGAUGCUCA --------------((-(((...(((..(((((((.((((.....)))).((((.(((...))).)))).....)))).)))...))).)))))..................... ( -20.60) >DroPse_CAF1 151795 98 - 1 --U---------------GUCACUUACUGCGUGACAAAUUCCUUGGCUUCGAUGGCAAUUCUCCCAGCCAAAUCGUGAAUAUUUAUGAUGAGCUCCACUACGGAAUUCAUCUGGA --(---------------(((((.......))))))...(((((((((..((.((....))))..))))))...............((((((.(((.....))).)))))).))) ( -25.30) >DroSec_CAF1 143450 112 - 1 ---UCCACAAAUAAAGCAAUUACUCACUGCGACACAAACGCCCUGGCUUCGUUCGCCACUCUAGAAGCCAAAUCGUGUAUGUUUAUGACCAGAUCCACAAUGGAUCUUAUCUGAA ---............(((.........)))((((...(((...(((((((.............)))))))...)))...))))...((..((((((.....))))))..)).... ( -24.82) >DroEre_CAF1 152740 113 - 1 --UUCCACAAAUAAGGCUUCUACUUACUGGGAUACAAAUCCCCUGGCUUCGUUGGCCACUCUAGAAGCCAAAUCAUGUAUGUUUAUGACCAGAUCCACAAUGGAUCGUAUCUGGA --.((((.......((((((((......(((((....))))).(((((.....)))))...))))))))...(((((......)))))...(((((.....))))).....)))) ( -37.40) >DroYak_CAF1 148934 113 - 1 --UUUCACAAAUAAAUCAUUUACUCACUGGGACACAAAUCCCCUAGCUUCGUUGGCCACUCUAGAAGCCAAAUCGUGUAUGUUUAUGACCAGAUCCACAAUGGAUCGUAUCUGGA --.............((((..((.(((.((((......))))...(((((...(....)....)))))......)))...))..))))((((((((.....))).....))))). ( -25.50) >consensus __UUCCACAAAUAAAGCAUUUACUCACUGCGACACAAAUCCCCUGGCUUCGUUCGCCACUCUAGAAGCCAAAUCGUGUAUGUUUAUGACCAGAUCCACAAUGGAUCUUAUCUGGA ...........................................(((((((.............)))))))..(((((......)))))...(((((.....)))))......... (-12.03 = -13.04 + 1.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:26:50 2006