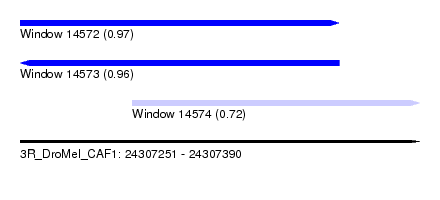
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 24,307,251 – 24,307,390 |
| Length | 139 |
| Max. P | 0.970498 |

| Location | 24,307,251 – 24,307,362 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 78.87 |
| Mean single sequence MFE | -33.62 |
| Consensus MFE | -19.05 |
| Energy contribution | -18.91 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.80 |
| Structure conservation index | 0.57 |
| SVM decision value | 1.66 |
| SVM RNA-class probability | 0.970498 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
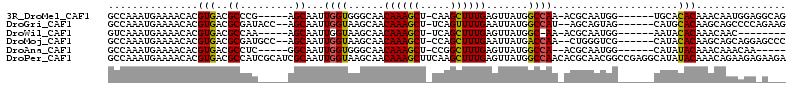
>3R_DroMel_CAF1 24307251 111 + 27905053 G-AGCACUUGGCACAAGGCGUCCUUGCGAGAACGAGAGGCGCCAAAUGAAAACACGUGACGCCCG-----AGCAAUUGGUGGGCAACAAAGCU-CAAGCUUUGAGUUAUGGCCAA-ACG .-.....(((((....((((((((((......)))).))))))...........(((((((((((-----..(....).)))))..((((((.-...)))))).)))))))))))-... ( -43.70) >DroVir_CAF1 110625 99 + 1 A-AGCACUUGGCAUGAG-------------AGCG-AAGAAGCCAAAUGAAAACACGUGACGCGAUGCC--AGCAAUUGGUAAGCAACAAAGCU-UCAGCUUUGAGUUAUGGCCAA--CC .-.....(((((....(-------------.((.-.....)))...........((((((((..((((--((...)))))).))..((((((.-...)))))).)))))))))))--.. ( -28.90) >DroPse_CAF1 136238 111 + 1 GAAGCACUUGGCAAAAAG-------GCGAGAGCG-GAGGCGCCAAAUGAAAACACGUGACGCCAUCGCAUCGCAAUUGGUAAGCAACAAAGCUUCAAGCUUUGAGUUAUGGCCAACACG .......(((((......-------((((..(((-..((((.((..((....))..)).))))..))).)))).............((((((.....)))))).......))))).... ( -35.50) >DroWil_CAF1 101814 96 + 1 A-AGCACUUGGCAUAA--------------AACGAAAGGCGUCAAAUGAAAACACGUGACGCCAA-----AGCAAUUGGUAAGCAACAAAGCU-UCAGCUUUGAGUUAUGGC-AA-ACG .-.....(((.(((((--------------.......(((((((..((....))..)))))))..-----................((((((.-...))))))..))))).)-))-... ( -27.90) >DroAna_CAF1 132743 96 + 1 A-AGCACUUGGCAUAAAGC--------------AAGAGGCGCCAAAUGAAAACACGUGACGCCUC-----GGCAAUUGGUGGGCAACAAAGCU-CCGGCUUUGAGUUAUGGCCA--ACG .-.....(((((((((..(--------------((((((((.((..((....))..)).))))))-----.......(.(((((......)).-))).).)))..)))).))))--).. ( -30.20) >DroPer_CAF1 136024 111 + 1 GAAGCACUUGGCAAAAAG-------GCGAGAGCG-GAGGCGCCAAAUGAAAACACGUGACGCCAUCGCAUCGCAAUUGGUAAGCAACAAAGCUUCAAGCUUUGAGUUAUGGCCAACACG .......(((((......-------((((..(((-..((((.((..((....))..)).))))..))).)))).............((((((.....)))))).......))))).... ( -35.50) >consensus A_AGCACUUGGCAUAAAG____________AGCG_GAGGCGCCAAAUGAAAACACGUGACGCCAU_____AGCAAUUGGUAAGCAACAAAGCU_CAAGCUUUGAGUUAUGGCCAA_ACG ...((..(((((............................))))).........((((((((.........)).............((((((.....)))))).))))))))....... (-19.05 = -18.91 + -0.14)


| Location | 24,307,251 – 24,307,362 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 78.87 |
| Mean single sequence MFE | -33.01 |
| Consensus MFE | -23.21 |
| Energy contribution | -23.60 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.70 |
| SVM decision value | 1.54 |
| SVM RNA-class probability | 0.962295 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24307251 111 - 27905053 CGU-UUGGCCAUAACUCAAAGCUUG-AGCUUUGUUGCCCACCAAUUGCU-----CGGGCGUCACGUGUUUUCAUUUGGCGCCUCUCGUUCUCGCAAGGACGCCUUGUGCCAAGUGCU-C ((.-.((((.......((((((...-.))))))..((((..........-----.))))))))..))....((((((((((....((((((....))))))....))))))))))..-. ( -36.40) >DroVir_CAF1 110625 99 - 1 GG--UUGGCCAUAACUCAAAGCUGA-AGCUUUGUUGCUUACCAAUUGCU--GGCAUCGCGUCACGUGUUUUCAUUUGGCUUCUU-CGCU-------------CUCAUGCCAAGUGCU-U ((--(.(((.......((((((...-.))))))..))).)))....(((--(((.....)))).)).....((((((((.....-....-------------.....))))))))..-. ( -24.94) >DroPse_CAF1 136238 111 - 1 CGUGUUGGCCAUAACUCAAAGCUUGAAGCUUUGUUGCUUACCAAUUGCGAUGCGAUGGCGUCACGUGUUUUCAUUUGGCGCCUC-CGCUCUCGC-------CUUUUUGCCAAGUGCUUC .((((((((.......((((((.....)))))).............((((.(((..(((((((.(((....))).)))))))..-)))..))))-------......))))).)))... ( -38.30) >DroWil_CAF1 101814 96 - 1 CGU-UU-GCCAUAACUCAAAGCUGA-AGCUUUGUUGCUUACCAAUUGCU-----UUGGCGUCACGUGUUUUCAUUUGACGCCUUUCGUU--------------UUAUGCCAAGUGCU-U (((-((-(.(((((..((((((..(-(((......)))).......)))-----))(((((((.(((....))).)))))))....)..--------------))))).))))))..-. ( -27.40) >DroAna_CAF1 132743 96 - 1 CGU--UGGCCAUAACUCAAAGCCGG-AGCUUUGUUGCCCACCAAUUGCC-----GAGGCGUCACGUGUUUUCAUUUGGCGCCUCUU--------------GCUUUAUGCCAAGUGCU-U ..(--((((.((((..(((....((-.((......))))..........-----(((((((((.(((....))).)))))))))))--------------)..))))))))).....-. ( -32.70) >DroPer_CAF1 136024 111 - 1 CGUGUUGGCCAUAACUCAAAGCUUGAAGCUUUGUUGCUUACCAAUUGCGAUGCGAUGGCGUCACGUGUUUUCAUUUGGCGCCUC-CGCUCUCGC-------CUUUUUGCCAAGUGCUUC .((((((((.......((((((.....)))))).............((((.(((..(((((((.(((....))).)))))))..-)))..))))-------......))))).)))... ( -38.30) >consensus CGU_UUGGCCAUAACUCAAAGCUGG_AGCUUUGUUGCUUACCAAUUGCU_____AUGGCGUCACGUGUUUUCAUUUGGCGCCUC_CGCU____________CUUUAUGCCAAGUGCU_C ....(((((.......((((((.....)))))).......................(((((((.(((....))).))))))).........................)))))....... (-23.21 = -23.60 + 0.39)

| Location | 24,307,290 – 24,307,390 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 74.56 |
| Mean single sequence MFE | -25.61 |
| Consensus MFE | -12.73 |
| Energy contribution | -12.40 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.50 |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.716261 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24307290 100 + 27905053 GCCAAAUGAAAACACGUGACGCCCG-----AGCAAUUGGUGGGCAACAAAGCU-CAAGCUUUGAGUUAUGGCCAA-ACGCAAUGG------UGCACACAAACAAUGGAGGCAG (((...........(((((((((((-----..(....).)))))..((((((.-...)))))).))))))((((.-......)))------)................))).. ( -30.10) >DroGri_CAF1 112206 102 + 1 GCCAAAUGAAAACACGUGACGCGAUACC--AGCAAUUGGUAAGCAACAAAGCU-UCAGUUUUGAAUUAUGGCCAU--AGCAGUAG------CAUGCACAAGCAGCCCCAGAAG ((((................((..((((--((...)))))).))..((((((.-...)))))).....))))...--....(..(------(.(((....)))))..)..... ( -22.20) >DroWil_CAF1 101839 91 + 1 GUCAAAUGAAAACACGUGACGCCAA-----AGCAAUUGGUAAGCAACAAAGCU-UCAGCUUUGAGUUAUGGC-AA-ACGCAAUGG------AAUACACAAACAAC-------- ((((..((....))..))))((((.-----(((..(((.....)))((((((.-...)))))).))).))))-..-.........------..............-------- ( -20.40) >DroMoj_CAF1 89549 102 + 1 GCCAAAUGAAAACACGUGACGCGAUGCC--AGCAAUUGGUAAGCAACAAAGCU-CCAGCUUUGAAUUAUGACCAA--CUGGGUCG------CAUACACAAGCAGCAGGAGCCC (((...((.......(((..(((((.((--((...(((((......((((((.-...)))))).......)))))--))))))))------)...)))......)).).)).. ( -26.64) >DroAna_CAF1 132768 94 + 1 GCCAAAUGAAAACACGUGACGCCUC-----GGCAAUUGGUGGGCAACAAAGCU-CCGGCUUUGAGUUAUGGCCA--ACGCAAUGG------CAUAUACAAACAAACAA----- ((((..........(((((((((.(-----(.(....).)))))..((((((.-...)))))).))))))((..--..))..)))------)................----- ( -27.30) >DroPer_CAF1 136056 113 + 1 GCCAAAUGAAAACACGUGACGCCAUCGCAUCGCAAUUGGUAAGCAACAAAGCUUCAAGCUUUGAGUUAUGGCCAACACGCAACGGCCGAGGCAUAUACAAACAGAAGAGAAGA (((............(((((((((..((...))...))))......((((((.....)))))).)))))((((..........))))..)))..................... ( -27.00) >consensus GCCAAAUGAAAACACGUGACGCCAU_____AGCAAUUGGUAAGCAACAAAGCU_CCAGCUUUGAGUUAUGGCCAA_ACGCAAUGG______CAUACACAAACAACAGA_G___ ...............(((..((.........))...((((......((((((.....)))))).......)))).....................)))............... (-12.73 = -12.40 + -0.33)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:26:43 2006