
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 24,274,949 – 24,275,049 |
| Length | 100 |
| Max. P | 0.940709 |

| Location | 24,274,949 – 24,275,049 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 78.25 |
| Mean single sequence MFE | -31.49 |
| Consensus MFE | -18.66 |
| Energy contribution | -19.22 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.59 |
| SVM decision value | 1.31 |
| SVM RNA-class probability | 0.940709 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
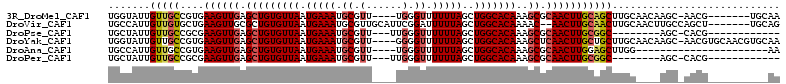
>3R_DroMel_CAF1 24274949 100 + 27905053 UGGUAUUGUUGCCGUGAAGUUGAGCUGUGUUAAUGAAAUGCGUU----UGGGUUUUUUAGCUGGCACAAAGCGCAACUUGCAGCUUGCAACAAGC-AACG-------UGCAA .((((....))))...((((((.(((((((((.(((((.((...----...)).)))))..))))))..))).))))))(((..((((.....))-))..-------))).. ( -29.50) >DroVir_CAF1 74237 103 + 1 UGCCAUUGUUGUGCUGAAGUUGCGCUGUGUUAAUGAAAUGCGUUGCAUUCGGAUUUUUAGCUGGCACAAAAC--AACUUGCAACUUGCAACUUGCCAGCU-------UGCAG (((..........(((((((.((((..............)))).)).)))))......((((((((......--...((((.....))))..))))))))-------.))). ( -26.54) >DroPse_CAF1 101131 88 + 1 UGCUAUUGUUGCCGCGAAGUUGAGCUGUGUUAAUGAAAUGCGUU---UUGGGUUUUUUAGCUGGCACAAAGCGCAACUUGCGGC--------AGC-CACG------------ .......((((((((.((((((.(((((((((.(((((.((...---....)).)))))..))))))..))).)))))))))))--------)))-....------------ ( -34.10) >DroYak_CAF1 102003 107 + 1 UGGUAUUGUUGCCGUGAAGUUGAGCUGUGUUAAUGAAAUGCGUU----GGGGUUUUUUAGCUGGCACAAAGCUCAACUUGCUGCUUGCAACAAGC-AACGUGCAACGUGCAA ..((((.((((((((.((((((((((((((((.(((((.((...----...)).)))))..))))))..))))))))))..((((((...)))))-)))).))))))))).. ( -40.90) >DroAna_CAF1 95124 86 + 1 UGCCAUUGUUGCCGUGAAGUUGAGCUGUGUUAAUGAAAUGCGUU----UGGGUUUUUUAGCUGGCACAAAGCGCAACUUGGAGCUUGG----------------------AA ..(((..(((.(((...(((((.(((((((((.(((((.((...----...)).)))))..))))))..))).))))))))))).)))----------------------.. ( -23.80) >DroPer_CAF1 101005 88 + 1 UGCUAUUGUUGCCGCGAAGUUGAGCUGUGUUAAUGAAAUGCGUU---UUGGGUUUUUUAGCUGGCACAAAGCGCAACUUGCGGC--------AGC-CACG------------ .......((((((((.((((((.(((((((((.(((((.((...---....)).)))))..))))))..))).)))))))))))--------)))-....------------ ( -34.10) >consensus UGCUAUUGUUGCCGUGAAGUUGAGCUGUGUUAAUGAAAUGCGUU____UGGGUUUUUUAGCUGGCACAAAGCGCAACUUGCAGCUUGC____AGC_AACG__________AA .......(((((....((((((.(((((((((.(((((.((.(......).)).)))))..)))))))..)).)))))))))))............................ (-18.66 = -19.22 + 0.56)


| Location | 24,274,949 – 24,275,049 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 78.25 |
| Mean single sequence MFE | -23.51 |
| Consensus MFE | -12.21 |
| Energy contribution | -12.52 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.52 |
| SVM decision value | 0.71 |
| SVM RNA-class probability | 0.828810 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24274949 100 - 27905053 UUGCA-------CGUU-GCUUGUUGCAAGCUGCAAGUUGCGCUUUGUGCCAGCUAAAAAACCCA----AACGCAUUUCAUUAACACAGCUCAACUUCACGGCAACAAUACCA ..((.-------....-))(((((((....((.((((((.(((.(((.................----..............))).))).))))))))..)))))))..... ( -20.68) >DroVir_CAF1 74237 103 - 1 CUGCA-------AGCUGGCAAGUUGCAAGUUGCAAGUU--GUUUUGUGCCAGCUAAAAAUCCGAAUGCAACGCAUUUCAUUAACACAGCGCAACUUCAGCACAACAAUGGCA .(((.-------((((((((....((((.(....).))--))....))))))))........((((((...))))))............)))......((.((....)))). ( -25.60) >DroPse_CAF1 101131 88 - 1 ------------CGUG-GCU--------GCCGCAAGUUGCGCUUUGUGCCAGCUAAAAAACCCAA---AACGCAUUUCAUUAACACAGCUCAACUUCGCGGCAACAAUAGCA ------------.((.-(.(--------(((((((((((.(((.(((..................---..............))).))).)))))).)))))).)....)). ( -24.45) >DroYak_CAF1 102003 107 - 1 UUGCACGUUGCACGUU-GCUUGUUGCAAGCAGCAAGUUGAGCUUUGUGCCAGCUAAAAAACCCC----AACGCAUUUCAUUAACACAGCUCAACUUCACGGCAACAAUACCA ......(((((..(((-(((((...))))))))((((((((((.(((.................----..............))).))))))))))....)))))....... ( -32.08) >DroAna_CAF1 95124 86 - 1 UU----------------------CCAAGCUCCAAGUUGCGCUUUGUGCCAGCUAAAAAACCCA----AACGCAUUUCAUUAACACAGCUCAACUUCACGGCAACAAUGGCA ..----------------------....(((..((((((.(((.(((.................----..............))).))).))))))...(....)...))). ( -13.78) >DroPer_CAF1 101005 88 - 1 ------------CGUG-GCU--------GCCGCAAGUUGCGCUUUGUGCCAGCUAAAAAACCCAA---AACGCAUUUCAUUAACACAGCUCAACUUCGCGGCAACAAUAGCA ------------.((.-(.(--------(((((((((((.(((.(((..................---..............))).))).)))))).)))))).)....)). ( -24.45) >consensus UU__________CGUU_GCU____GCAAGCUGCAAGUUGCGCUUUGUGCCAGCUAAAAAACCCA____AACGCAUUUCAUUAACACAGCUCAACUUCACGGCAACAAUAGCA ............................((((..(((((.(((.(((...................................))).))).)))))...)))).......... (-12.21 = -12.52 + 0.31)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:26:29 2006