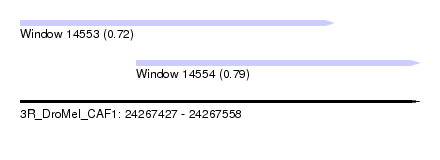
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 24,267,427 – 24,267,558 |
| Length | 131 |
| Max. P | 0.791737 |

| Location | 24,267,427 – 24,267,530 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 82.89 |
| Mean single sequence MFE | -43.00 |
| Consensus MFE | -29.33 |
| Energy contribution | -30.63 |
| Covariance contribution | 1.31 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.723884 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
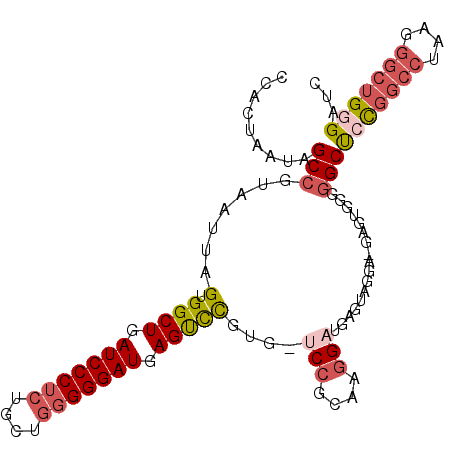
>3R_DroMel_CAF1 24267427 103 + 27905053 CCACUAAUAGCCGUAAUUAGUGGCUGAUCCCUCUGCUGGGGGAUGAGUCCGUG-UCCGCAAGGAUGAGUCGGA-GAGUGGGGGCUCCGGCCUAAGGGCUGGGCUU (((((.....(((......(.((((.(((((((....))))))).)))))..(-(((....))))....))).-.)))))((((.((((((....)))))))))) ( -50.80) >DroSec_CAF1 90197 103 + 1 CCACUAAUAGCCGUAAUUAGUGGCUGAUCCCUCUGCUGGGGGAUGAGUCCGUG-UCCGCAAGGAUGAGUUGGA-GAGUGGGGGCUCCGGCCUAAGGGCUGGGAUC (((((..(((((.......(.((((.(((((((....))))))).)))))..(-(((....))))).))))..-.))))).((..((((((....))))))..)) ( -47.40) >DroSim_CAF1 94887 103 + 1 CCACUAAUAGCCGUAAUUAGUGGCUGAUCCCUCUGCUGGGGGAUGAGUCCGUG-UCCGCAAGGCUGAGUUGGA-GAGUGGGGGCUCUGGCCUAAGGGCUGGGAUC (((((.....(((...(((((((((.(((((((....))))))).))))....-..(....))))))..))).-.))))).((..(..(((....)))..)..)) ( -42.60) >DroEre_CAF1 97452 103 + 1 CCACUAAUAGCCGUAAUUAGUGGCUGAUCCCUCAGCUGGGGGAUGAGUUCGCG-UCCGCAGGGAUGAGUAGGU-GGGCUUGGGCUCCGGCCUAAGGGCUGGGAUC ........((((.......((((((.(((((((....))))))).)).))))(-(((((..(......)..))-))))...))))((((((....)))))).... ( -42.80) >DroYak_CAF1 94321 104 + 1 CCACUAAUAGCCGUAAUUAGUGGCUGAUCCCGCUGCUGGGGGAUGAGUCCGUG-UCCGCAAGGAUGAGUAGGUUGGGAUUGGGCUACGGCCCAAGGGCUGGGAUC ((((((((.......))))))))..((((((.(((((...(((....)))..(-(((....)))).)))))....((.((((((....))))))...)))))))) ( -45.50) >DroAna_CAF1 87636 92 + 1 CCACUAAUAGCCGUAAUUAGUGGCUGAUCCCU-UGACGGGGGAUGAGCCUACCCCCCGCA--GCUGAGU--------CUUGAGCCCCACC--AGCUCCCGAGCUC ........(((.((.....((((((.((((((-.....)))))).)))).)).....)).--)))(((.--------(((((((......--.))))..)))))) ( -28.90) >consensus CCACUAAUAGCCGUAAUUAGUGGCUGAUCCCUCUGCUGGGGGAUGAGUCCGUG_UCCGCAAGGAUGAGUAGGA_GAGUGGGGGCUCCGGCCUAAGGGCUGGGAUC .........(((.......(.((((.(((((((....))))))).)))))....(((....))).................)))(((((((....)))))))... (-29.33 = -30.63 + 1.31)

| Location | 24,267,465 – 24,267,558 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 88.03 |
| Mean single sequence MFE | -38.02 |
| Consensus MFE | -27.20 |
| Energy contribution | -27.12 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.59 |
| SVM RNA-class probability | 0.791737 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
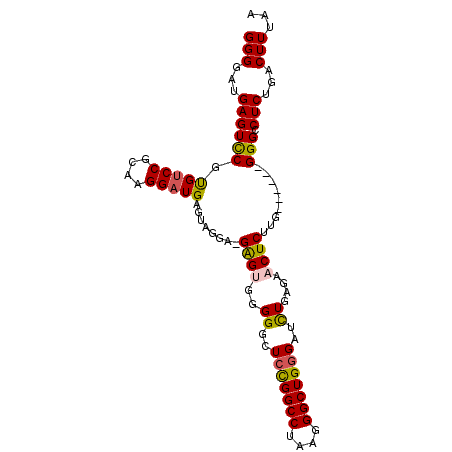
>3R_DroMel_CAF1 24267465 93 + 27905053 GGGGAUGAGUCCGUGUCCGCAAGGAUGAGUCGGA-GAGUGGGGGCUCCGGCCUAAGGGCUGGGCUUUGAGAACUCUUG------GGGCCUCUGACUUUAA ..(((....)))..((((....))))((((((((-(..........((((((....))))))((((..((....))..------)))))))))))))... ( -41.50) >DroSec_CAF1 90235 93 + 1 GGGGAUGAGUCCGUGUCCGCAAGGAUGAGUUGGA-GAGUGGGGGCUCCGGCCUAAGGGCUGGGAUCUGAGAACUCUUG------GGGCCUCUGACUUUAA ..(((....)))..((((....)))).......(-(((((((((((((((((....))))))).((..((....))..------)))))))).))))).. ( -38.30) >DroSim_CAF1 94925 93 + 1 GGGGAUGAGUCCGUGUCCGCAAGGCUGAGUUGGA-GAGUGGGGGCUCUGGCCUAAGGGCUGGGAUCUGAGAACUCUUG------GGGCCUCUGACUUUAA ..(((....)))..(.((....)))........(-((((((((((((..(((....)))..)).((..((....))..------)))))))).))))).. ( -32.30) >DroEre_CAF1 97490 99 + 1 GGGGAUGAGUUCGCGUCCGCAGGGAUGAGUAGGU-GGGCUUGGGCUCCGGCCUAAGGGCUGGGAUCUGAGAACUCCUGGGGUUGGGGGCUCUGACUUUAA (((...((((((.((.((.(((((..........-(..((..((..((((((....))))))..))..))..)))))).)).)).))))))...)))... ( -41.20) >DroYak_CAF1 94359 95 + 1 GGGGAUGAGUCCGUGUCCGCAAGGAUGAGUAGGUUGGGAUUGGGCUACGGCCCAAGGGCUGGGAUCUGAGAACUCUUGG-----GGGGCUCUGACUUUAA (((...((((((..((((....))))(((((((((.((.((((((....))))))...)).).))))....))))....-----.))))))...)))... ( -36.80) >consensus GGGGAUGAGUCCGUGUCCGCAAGGAUGAGUAGGA_GAGUGGGGGCUCCGGCCUAAGGGCUGGGAUCUGAGAACUCUUG______GGGCCUCUGACUUUAA (((...((((((.(((((....)))))........((((..((..(((((((....)))))))..))....)))).........))).)))...)))... (-27.20 = -27.12 + -0.08)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:26:26 2006