
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 24,266,928 – 24,267,031 |
| Length | 103 |
| Max. P | 0.889067 |

| Location | 24,266,928 – 24,267,031 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 79.12 |
| Mean single sequence MFE | -30.02 |
| Consensus MFE | -17.59 |
| Energy contribution | -17.03 |
| Covariance contribution | -0.55 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.611554 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
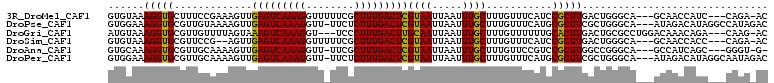
>3R_DroMel_CAF1 24266928 103 + 27905053 GUGUAAAGGUGCUUUCCGAAAGUUGAGUCAAAGGUUUUUCGCUUUGACUCGUAAUUAAUUUGCUUUGUUUCAUCCGCUUGACUGGGCA---GCAACCAUC---CAGA-AC .......((((((.((((.((((.(((((((((........)))))))))((((.....))))............))))...)))).)---)).)))...---....-.. ( -27.50) >DroPse_CAF1 92497 106 + 1 GUGGAAAGGUGCGUUGUAAAAGUUGAGUCAAAGGUU-UUCUCUUUGACUCGUAAUUAAUUUGCUUUGUUUCAUGCGCUUCGCUGGGCA---AUAGACAUAGGCCAUAGAC ..((..((((((((....((((((((((((((((..-...)))))))))))..........))))).....))))))))..)).(((.---..........)))...... ( -28.20) >DroGri_CAF1 71234 103 + 1 AUGUAAAGGUGCGUUGUUUUAGUAAAGUCAAAGGU---UCCCUUUGACUUGCAAUUAAUUUGCUUUGUUUUUUGCACUUGACUGCGCCUGGACAAACAGA---CAAG-AC .(((..(((((((..(((..(((.((((((((((.---..))))))))))((((..(((.......)))..))))))).))))))))))..)))......---....-.. ( -32.20) >DroSim_CAF1 94399 101 + 1 GUGUAAAGGUGCGUUCCG--AGUUGAGUCAAAGGUUUUUCGCUUUGACUCGUAAUUAAUUUGCUUUGUUUCAUCCGCUUGACUGGGCA---GCAACCACC---CAGA-AC .......(((((...((.--(((((((((((((........)))))))))((((.....))))................)))).))..---)).)))...---....-.. ( -27.60) >DroAna_CAF1 87136 101 + 1 GUGCAAAGGUGCGUUGCAAAAGUUGAGUCAAAGGUU-UUCGCUUUGACUCGUAAUUAAUUUGCUUUGUUCCGUCCGCUUGGCCGGGCA---GCCAUCAGC---GGGU-G- .......((.(((..(((((...((((((((((...-....)))))))))).......)))))..))).)).((((((((((......---))))..)))---))).-.- ( -35.60) >DroPer_CAF1 92297 106 + 1 GUGGAAAGGUGCGUUGCAAAAGUUGAGUCAAAGGUU-UUCUCUUUGACUCGUAAUUAAUUUGCUUUGUUUCAUGCGCUUCGCUGGGCA---AUAGACAUAGGCAAUAGAC ......((((((((.(((((....((((((((((..-...))))))))))((((.....)))))))))...)))))))).(((.(...---.....)...)))....... ( -29.00) >consensus GUGUAAAGGUGCGUUGCAAAAGUUGAGUCAAAGGUU_UUCGCUUUGACUCGUAAUUAAUUUGCUUUGUUUCAUCCGCUUGACUGGGCA___ACAAACAUA___CAGA_AC ......(((((.............(((((((((........)))))))))((((.....))))...........)))))............................... (-17.59 = -17.03 + -0.55)
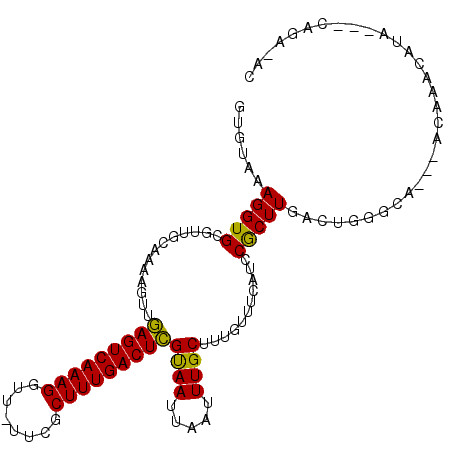
| Location | 24,266,928 – 24,267,031 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 79.12 |
| Mean single sequence MFE | -26.08 |
| Consensus MFE | -14.80 |
| Energy contribution | -14.38 |
| Covariance contribution | -0.41 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.95 |
| SVM RNA-class probability | 0.889067 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
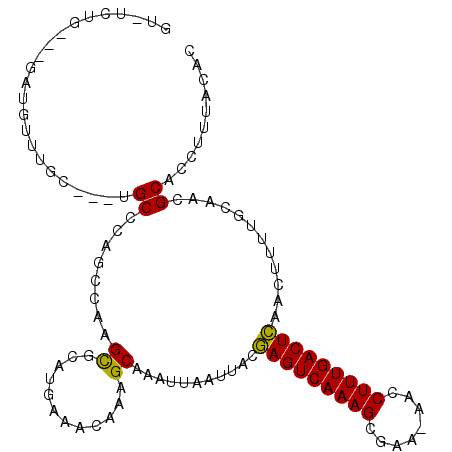
>3R_DroMel_CAF1 24266928 103 - 27905053 GU-UCUG---GAUGGUUGC---UGCCCAGUCAAGCGGAUGAAACAAAGCAAAUUAAUUACGAGUCAAAGCGAAAAACCUUUGACUCAACUUUCGGAAAGCACCUUUACAC .(-((((---((.((((((---(......(((......))).....)))...........(((((((((........))))))))))))))))))))............. ( -24.50) >DroPse_CAF1 92497 106 - 1 GUCUAUGGCCUAUGUCUAU---UGCCCAGCGAAGCGCAUGAAACAAAGCAAAUUAAUUACGAGUCAAAGAGAA-AACCUUUGACUCAACUUUUACAACGCACCUUUCCAC ......(((..........---.)))..(((..((............))...........(((((((((....-...)))))))))...........))).......... ( -19.90) >DroGri_CAF1 71234 103 - 1 GU-CUUG---UCUGUUUGUCCAGGCGCAGUCAAGUGCAAAAAACAAAGCAAAUUAAUUGCAAGUCAAAGGGA---ACCUUUGACUUUACUAAAACAACGCACCUUUACAU (.-((((---((((......)))))).)).)..((((..........((((.....))))((((((((((..---.))))))))))............))))........ ( -27.40) >DroSim_CAF1 94399 101 - 1 GU-UCUG---GGUGGUUGC---UGCCCAGUCAAGCGGAUGAAACAAAGCAAAUUAAUUACGAGUCAAAGCGAAAAACCUUUGACUCAACU--CGGAACGCACCUUUACAC ((-((((---(((..((((---(......(((......))).....))))).........(((((((((........))))))))).)))--))))))............ ( -30.70) >DroAna_CAF1 87136 101 - 1 -C-ACCC---GCUGAUGGC---UGCCCGGCCAAGCGGACGGAACAAAGCAAAUUAAUUACGAGUCAAAGCGAA-AACCUUUGACUCAACUUUUGCAACGCACCUUUGCAC -.-..((---(((..((((---(....))))))))))..........(((((........(((((((((.(..-..))))))))))......(((...)))..))))).. ( -32.20) >DroPer_CAF1 92297 106 - 1 GUCUAUUGCCUAUGUCUAU---UGCCCAGCGAAGCGCAUGAAACAAAGCAAAUUAAUUACGAGUCAAAGAGAA-AACCUUUGACUCAACUUUUGCAACGCACCUUUCCAC ......(((.(((((((.(---(((...)))))).))))).......(((((........(((((((((....-...)))))))))....)))))...)))......... ( -21.80) >consensus GU_UCUG___GAUGUUUGC___UGCCCAGCCAAGCGCAUGAAACAAAGCAAAUUAAUUACGAGUCAAAGCGAA_AACCUUUGACUCAACUUUUGCAACGCACCUUUACAC .......................((........((............))...........(((((((((........)))))))))............)).......... (-14.80 = -14.38 + -0.41)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:26:23 2006