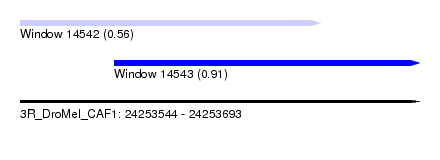
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 24,253,544 – 24,253,693 |
| Length | 149 |
| Max. P | 0.913702 |

| Location | 24,253,544 – 24,253,656 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 89.41 |
| Mean single sequence MFE | -41.83 |
| Consensus MFE | -32.35 |
| Energy contribution | -33.43 |
| Covariance contribution | 1.09 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.56 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.561771 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
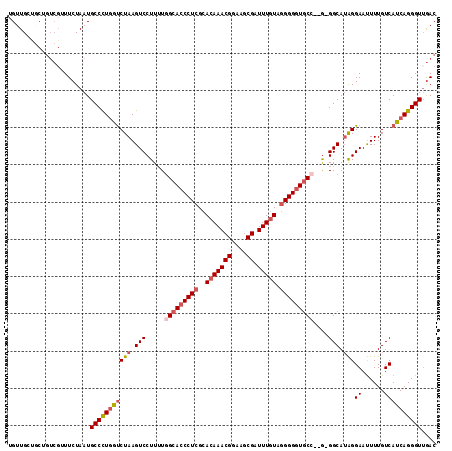
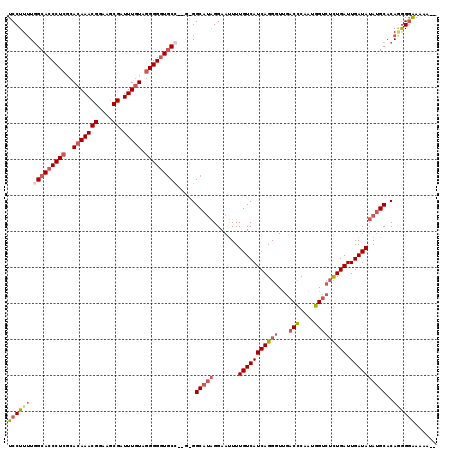
>3R_DroMel_CAF1 24253544 112 + 27905053 UGUUGCUGCUGUCGUUUCUAAUGCCCUGAUCUAAGUCCUUUUGGCACCCUCGCACAAACGGAAGCGAUUUGUAGGGGGUGCC--G-GGCAAAGGAAUUUUGUCAUCAGGGUUGAC ..........((((........((((((((.(((.((((((((((((((((..(((((((....)).))))).)))))))))--)-...))))))...)))..)))))))))))) ( -43.40) >DroSec_CAF1 76508 112 + 1 UGUUGCUGCUGUCGUUUCUAAUGCCCUGGUCUAAGUCCUUUUGGCACCCUCGCACAAACGGAAGCGAUUUGUAGGGGGUGCC--G-GGCAUAGGAAUUUUGUCAUCAGGGUAGAC .....(((.((.((.((((.((((((.((.......))....(((((((((..(((((((....)).))))).)))))))))--)-))))).))))...)).)).)))....... ( -45.70) >DroSim_CAF1 81055 112 + 1 UGUUGCUGCUGUCGCUUCUAAUGCCUUGGUCUAAGUCCUUUUGGCACCCUCGCACAAACGGAAGCGAUUUGUAGGGGGUGCC--G-GGCAUAGGAAUUUUGUCAUCAGGGUUGAC .....(((.((.((.((((.((((((.((.......))....(((((((((..(((((((....)).))))).)))))))))--)-))))).))))...)).)).)))....... ( -41.50) >DroEre_CAF1 81810 110 + 1 UGUUG---CUGUCGUUUCUAAUGCCCUGGUCUAAGUCCUUUUCGCACCCUCGCACAAACGGAAGCGAUUUGUAGGGGGUGCC--GGGGCAUAGGAAUUUUGUCAUCAGGGUUGAC .....---..((((........(((((((((((.(((((....((((((((..(((((((....)).))))).)))))))).--))))).)))((......)))))))))))))) ( -40.60) >DroYak_CAF1 80063 113 + 1 UGUUGCCGCUGUCGAUUCUAAUGCCCUAGUCUAAGUCCUUUUGGCACCCUCGCACAAACGGAAGCGAUUUGUAGGGGGUGCC--GGGGCAUAGGAAUUUUGUCAUCAGGGUUGAC ....(((.(((..((((((.(((((((.....(((....)))(((((((((..(((((((....)).))))).)))))))))--))))))).))).....)))..)))))).... ( -45.20) >DroAna_CAF1 73773 105 + 1 UGUUGC---------UUCUAAUGCCCUGGCCUAAGUCCUU-UGGCACCCUCGCACAAACGGAAACGAUUUUUGUGGGUUUCAAAAGGGCAUGGGAACUUUGUCAUCAGGGUUAAC ......---------.......(((((((((((.((((((-(((.((((..(((.(((((....)).))).))))))).)).))))))).))))..........))))))).... ( -34.60) >consensus UGUUGCUGCUGUCGUUUCUAAUGCCCUGGUCUAAGUCCUUUUGGCACCCUCGCACAAACGGAAGCGAUUUGUAGGGGGUGCC__G_GGCAUAGGAAUUUUGUCAUCAGGGUUGAC ......................(((((((((((.(((.....(((((((((..(((((((....)).))))).)))))))))....))).)))((......)))))))))).... (-32.35 = -33.43 + 1.09)


| Location | 24,253,579 – 24,253,693 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 88.84 |
| Mean single sequence MFE | -42.26 |
| Consensus MFE | -33.78 |
| Energy contribution | -35.39 |
| Covariance contribution | 1.62 |
| Combinations/Pair | 1.12 |
| Mean z-score | -3.14 |
| Structure conservation index | 0.80 |
| SVM decision value | 1.09 |
| SVM RNA-class probability | 0.913702 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
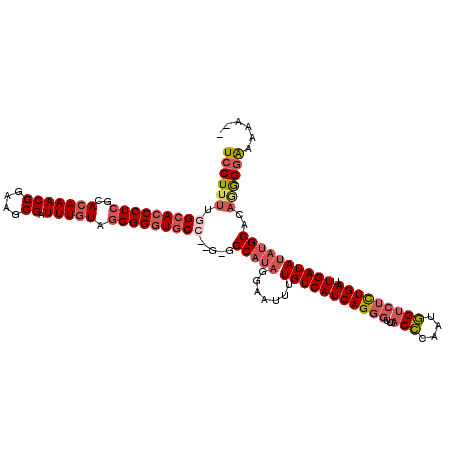
>3R_DroMel_CAF1 24253579 114 + 27905053 UCCUUUUGGCACCCUCGCACAAACGGAAGCGAUUUGUAGGGGGUGCC--G-GGCAAAGGAAUUUUGUCAUCAGGGUUGACCCAAUGGUCUCUGAUUGAUAUAUGCACAGGGGAAAAA-- ((((((((((((((((..(((((((....)).))))).)))))))))--)-...))))))....((((((((((...((((....))))))))).))))).................-- ( -43.10) >DroSec_CAF1 76543 114 + 1 UCCUUUUGGCACCCUCGCACAAACGGAAGCGAUUUGUAGGGGGUGCC--G-GGCAUAGGAAUUUUGUCAUCAGGGUAGACCCAUUGGUCUCUGAUUGAUAUAUGCACAGGGGAAAAA-- ((((((((((((((((..(((((((....)).))))).)))))))))--)-.(((((.......(((((((((...(((((....))))))))).))))))))))..))))))....-- ( -46.71) >DroSim_CAF1 81090 114 + 1 UCCUUUUGGCACCCUCGCACAAACGGAAGCGAUUUGUAGGGGGUGCC--G-GGCAUAGGAAUUUUGUCAUCAGGGUUGACCCAAUGGUCUCUGAUUGAUAUAUGCACAGGGGAAAAA-- ((((((((((((((((..(((((((....)).))))).)))))))))--)-.(((((.......((((((((((...((((....))))))))).))))))))))..))))))....-- ( -46.11) >DroEre_CAF1 81842 117 + 1 UCCUUUUCGCACCCUCGCACAAACGGAAGCGAUUUGUAGGGGGUGCC--GGGGCAUAGGAAUUUUGUCAUCAGGGUUGACCCAAUGGCCUCUGAUUGAUAUAUGCACAAGGGGAAAAAA (((((((.((((((((..(((((((....)).))))).)))))))).--...(((((.......(((((((((((((........)))).)))).))))))))))..)))))))..... ( -42.91) >DroYak_CAF1 80098 117 + 1 UCCUUUUGGCACCCUCGCACAAACGGAAGCGAUUUGUAGGGGGUGCC--GGGGCAUAGGAAUUUUGUCAUCAGGGUUGACCCAAUGGUCUCUGAUUGAUAUAUGCACAGGGGGAAAAAC ((((((((((((((((..(((((((....)).))))).)))))))))--...(((((.......((((((((((...((((....))))))))).))))))))))..)))))))..... ( -47.01) >DroAna_CAF1 73799 116 + 1 UCCUU-UGGCACCCUCGCACAAACGGAAACGAUUUUUGUGGGUUUCAAAAGGGCAUGGGAACUUUGUCAUCAGGGUUAACUCGAAAGU--UUGAUUGAUAAACGCACAUAGAAAAAAAC .((((-(((.((((..(((.(((((....)).))).))))))).)).)))))((.(((((((((((....)))))))..))))...((--(((.....))))))).............. ( -27.70) >consensus UCCUUUUGGCACCCUCGCACAAACGGAAGCGAUUUGUAGGGGGUGCC__G_GGCAUAGGAAUUUUGUCAUCAGGGUUGACCCAAUGGUCUCUGAUUGAUAUAUGCACAGGGGAAAAA__ ((((((.(((((((((..(((((((....)).))))).))))))))).....(((((.......(((((((((((...(((....))))))))).))))))))))..))))))...... (-33.78 = -35.39 + 1.62)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:26:16 2006