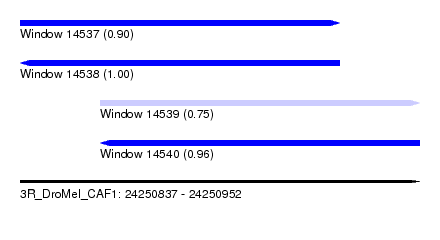
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 24,250,837 – 24,250,952 |
| Length | 115 |
| Max. P | 0.999626 |

| Location | 24,250,837 – 24,250,929 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 92 |
| Reading direction | forward |
| Mean pairwise identity | 89.87 |
| Mean single sequence MFE | -33.68 |
| Consensus MFE | -29.52 |
| Energy contribution | -29.92 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.02 |
| SVM RNA-class probability | 0.901796 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24250837 92 + 27905053 AGUUCGUCCUAGUCCUGCACCCGGAGUCCUGAGCCUGGCCUGAAGCACUUUUAGGCGCUCUUAGCGCGCCCAAACUUCGAGUGGCCAGGCUG .........(((..((.(....).))..)))((((((((((((((....(((.(((((.......))))).))))))))...))))))))). ( -34.80) >DroSec_CAF1 73765 90 + 1 AGUUCGUCCUAGUCCUGAACCUG--GUCCUGAGCCUGGCCUGAAGCACUUUUAGGCGCUCUUAGCGCGCCCAAACUUCGAGUGGCCAGGCUG .....(..((((........)))--)..)..((((((((((((((....(((.(((((.......))))).))))))))...))))))))). ( -34.40) >DroSim_CAF1 78307 92 + 1 AGUUCGUCCUAGUCCUGAACCCGGAGUCCUGAGCCUGGCCUGAAGCACUUUUAGGCGCUCUUAGCGCGCCCAAACUUCGAGUGGCCAGGCUG ((..(.(((..((.....))..))))..)).((((((((((((((....(((.(((((.......))))).))))))))...))))))))). ( -33.90) >DroEre_CAF1 79023 92 + 1 AGUUCGCCCUAGUCCUGAACACGGGGUCCUGAGCCUGGCCUGAAGCACUUUUAGGCGCCCUUAGCGCGCCCAAAGUUCGAGUGGCCAGGCUG .....(((((.((.......)))))))....(((((((((....(.(((((..(((((.......))))).))))).)....))))))))). ( -37.40) >DroYak_CAF1 77265 78 + 1 AGUUCACCCUAGUCC--------------UGAGCCUGGCCUGAAGCACUUUUAGGCGCUCUUAGCGCGCCCAAACUUCGAGUAGCCAGGCUG ...............--------------..((((((((((((((....(((.(((((.......))))).))))))).))..)))))))). ( -27.90) >consensus AGUUCGUCCUAGUCCUGAACCCGG_GUCCUGAGCCUGGCCUGAAGCACUUUUAGGCGCUCUUAGCGCGCCCAAACUUCGAGUGGCCAGGCUG ...............................((((((((((((((....(((.(((((.......))))).))))))))...))))))))). (-29.52 = -29.92 + 0.40)



| Location | 24,250,837 – 24,250,929 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 92 |
| Reading direction | reverse |
| Mean pairwise identity | 89.87 |
| Mean single sequence MFE | -38.50 |
| Consensus MFE | -34.24 |
| Energy contribution | -34.28 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.71 |
| Structure conservation index | 0.89 |
| SVM decision value | 3.80 |
| SVM RNA-class probability | 0.999626 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24250837 92 - 27905053 CAGCCUGGCCACUCGAAGUUUGGGCGCGCUAAGAGCGCCUAAAAGUGCUUCAGGCCAGGCUCAGGACUCCGGGUGCAGGACUAGGACGAACU .(((((((((....(((((((((((((.......))))))))....))))).))))))))).((..((((.(((.....))).))).)..)) ( -40.40) >DroSec_CAF1 73765 90 - 1 CAGCCUGGCCACUCGAAGUUUGGGCGCGCUAAGAGCGCCUAAAAGUGCUUCAGGCCAGGCUCAGGAC--CAGGUUCAGGACUAGGACGAACU .(((((((((....(((((((((((((.......))))))))....))))).))))))))).....(--(.((((...)))).))....... ( -38.90) >DroSim_CAF1 78307 92 - 1 CAGCCUGGCCACUCGAAGUUUGGGCGCGCUAAGAGCGCCUAAAAGUGCUUCAGGCCAGGCUCAGGACUCCGGGUUCAGGACUAGGACGAACU .(((((((((....(((((((((((((.......))))))))....))))).))))))))).((..((((.((((...)))).))).)..)) ( -40.40) >DroEre_CAF1 79023 92 - 1 CAGCCUGGCCACUCGAACUUUGGGCGCGCUAAGGGCGCCUAAAAGUGCUUCAGGCCAGGCUCAGGACCCCGUGUUCAGGACUAGGGCGAACU .(((((((((......(((((((((((.(...).)))))).)))))......))))))))).((..((((..(((...)))..))).)..)) ( -39.00) >DroYak_CAF1 77265 78 - 1 CAGCCUGGCUACUCGAAGUUUGGGCGCGCUAAGAGCGCCUAAAAGUGCUUCAGGCCAGGCUCA--------------GGACUAGGGUGAACU .(((((((((....(((((((((((((.......))))))))....))))).)))))))))..--------------............... ( -33.80) >consensus CAGCCUGGCCACUCGAAGUUUGGGCGCGCUAAGAGCGCCUAAAAGUGCUUCAGGCCAGGCUCAGGAC_CCGGGUUCAGGACUAGGACGAACU .(((((((((....(((((((((((((.......))))))))....))))).)))))))))............................... (-34.24 = -34.28 + 0.04)


| Location | 24,250,860 – 24,250,952 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 94 |
| Reading direction | forward |
| Mean pairwise identity | 82.51 |
| Mean single sequence MFE | -32.52 |
| Consensus MFE | -25.71 |
| Energy contribution | -26.68 |
| Covariance contribution | 0.97 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.749817 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24250860 92 + 27905053 GAGUCCUGAGCCUGGCCUGAAGCACUUUUAGGCGCUCUUAGCGCGCCCAAACUUCGAGUGGCCAGGCUGAAACGGCUAAA--ACGGCUGAAUGG ......(.((((((((((((((....(((.(((((.......))))).))))))))...))))))))).)..(((((...--..)))))..... ( -36.80) >DroSec_CAF1 73788 90 + 1 --GUCCUGAGCCUGGCCUGAAGCACUUUUAGGCGCUCUUAGCGCGCCCAAACUUCGAGUGGCCAGGCUGAAACAUUUAAA--ACGGCUGAAUGG --....(.((((((((((((((....(((.(((((.......))))).))))))))...))))))))).)..((((((..--.....)))))). ( -34.30) >DroSim_CAF1 78330 92 + 1 GAGUCCUGAGCCUGGCCUGAAGCACUUUUAGGCGCUCUUAGCGCGCCCAAACUUCGAGUGGCCAGGCUGAAACAGCUAAA--ACGGCUGAAUGG ......(.((((((((((((((....(((.(((((.......))))).))))))))...))))))))).)..(((((...--..)))))..... ( -37.50) >DroEre_CAF1 79046 92 + 1 GGGUCCUGAGCCUGGCCUGAAGCACUUUUAGGCGCCCUUAGCGCGCCCAAAGUUCGAGUGGCCAGGCUGAAACGGCUAAA--ACGGCUGAAUGG ......(.(((((((((....(.(((((..(((((.......))))).))))).)....))))))))).)..(((((...--..)))))..... ( -37.80) >DroYak_CAF1 77280 86 + 1 ------UGAGCCUGGCCUGAAGCACUUUUAGGCGCUCUUAGCGCGCCCAAACUUCGAGUAGCCAGGCUGAAACAGCUAAA--ACGACUGAAUGG ------(.((((((((((((((....(((.(((((.......))))).))))))).))..)))))))).)..((((....--..).)))..... ( -29.40) >DroAna_CAF1 70954 78 + 1 --GUCCUGAUCCCGGCCUGAAGCGAUUUUAUCC---------GCACACAAACUUCGAGUGCCCAGAAUGAACUGGAUGGAUGGCGGCUG----- --..........(((((....((.....(((((---------((((.(.......).))))((((......))))..))))))))))))----- ( -19.30) >consensus __GUCCUGAGCCUGGCCUGAAGCACUUUUAGGCGCUCUUAGCGCGCCCAAACUUCGAGUGGCCAGGCUGAAACAGCUAAA__ACGGCUGAAUGG ......(.((((((((((((((....(((.(((((.......))))).))))))))...))))))))).)..(((((.......)))))..... (-25.71 = -26.68 + 0.97)



| Location | 24,250,860 – 24,250,952 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 94 |
| Reading direction | reverse |
| Mean pairwise identity | 82.51 |
| Mean single sequence MFE | -34.47 |
| Consensus MFE | -30.17 |
| Energy contribution | -29.90 |
| Covariance contribution | -0.27 |
| Combinations/Pair | 1.27 |
| Mean z-score | -2.55 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.49 |
| SVM RNA-class probability | 0.958741 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24250860 92 - 27905053 CCAUUCAGCCGU--UUUAGCCGUUUCAGCCUGGCCACUCGAAGUUUGGGCGCGCUAAGAGCGCCUAAAAGUGCUUCAGGCCAGGCUCAGGACUC ............--.......((((.(((((((((....(((((((((((((.......))))))))....))))).)))))))))..)))).. ( -37.60) >DroSec_CAF1 73788 90 - 1 CCAUUCAGCCGU--UUUAAAUGUUUCAGCCUGGCCACUCGAAGUUUGGGCGCGCUAAGAGCGCCUAAAAGUGCUUCAGGCCAGGCUCAGGAC-- ............--.......((((.(((((((((....(((((((((((((.......))))))))....))))).)))))))))..))))-- ( -36.10) >DroSim_CAF1 78330 92 - 1 CCAUUCAGCCGU--UUUAGCUGUUUCAGCCUGGCCACUCGAAGUUUGGGCGCGCUAAGAGCGCCUAAAAGUGCUUCAGGCCAGGCUCAGGACUC ((...((((...--....))))....(((((((((....(((((((((((((.......))))))))....))))).)))))))))..)).... ( -40.80) >DroEre_CAF1 79046 92 - 1 CCAUUCAGCCGU--UUUAGCCGUUUCAGCCUGGCCACUCGAACUUUGGGCGCGCUAAGGGCGCCUAAAAGUGCUUCAGGCCAGGCUCAGGACCC ((....(((.((--....)).)))..(((((((((......(((((((((((.(...).)))))).)))))......)))))))))..)).... ( -36.30) >DroYak_CAF1 77280 86 - 1 CCAUUCAGUCGU--UUUAGCUGUUUCAGCCUGGCUACUCGAAGUUUGGGCGCGCUAAGAGCGCCUAAAAGUGCUUCAGGCCAGGCUCA------ .....((((...--....))))....(((((((((....(((((((((((((.......))))))))....))))).)))))))))..------ ( -35.50) >DroAna_CAF1 70954 78 - 1 -----CAGCCGCCAUCCAUCCAGUUCAUUCUGGGCACUCGAAGUUUGUGUGC---------GGAUAAAAUCGCUUCAGGCCGGGAUCAGGAC-- -----...((.((((((..((((......))))((((.(((...))).))))---------))))......((.....))..))....))..-- ( -20.50) >consensus CCAUUCAGCCGU__UUUAGCCGUUUCAGCCUGGCCACUCGAAGUUUGGGCGCGCUAAGAGCGCCUAAAAGUGCUUCAGGCCAGGCUCAGGAC__ ..........................(((((((((....(((((((((((((.......))))))))....))))).)))))))))........ (-30.17 = -29.90 + -0.27)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:26:13 2006