
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
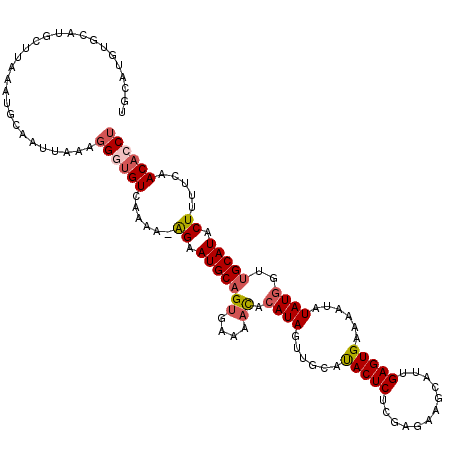
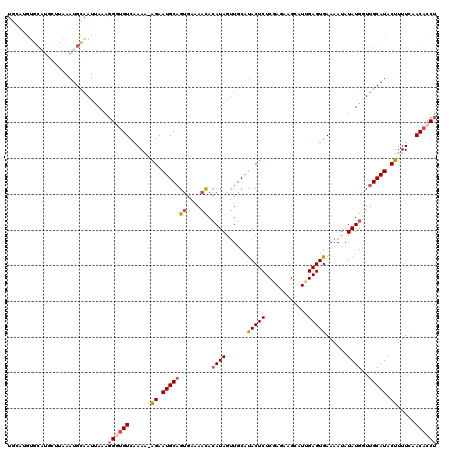
| Location | 24,239,590 – 24,239,709 |
| Length | 119 |
| Max. P | 0.949031 |

| Location | 24,239,590 – 24,239,709 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.56 |
| Mean single sequence MFE | -27.44 |
| Consensus MFE | -12.78 |
| Energy contribution | -14.74 |
| Covariance contribution | 1.96 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.47 |
| SVM decision value | 0.00 |
| SVM RNA-class probability | 0.535595 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24239590 119 + 27905053 UGCAUGUGCAUGCUUAAAUGCAAUUAAAGGGUGUCGAAA-AGAAUGCAGUGAAAACACAUAGUUGCAUACUCUCCAGAAGCAUUGAGUGAAAAUAUAUGGUUGCAUACUUUUCAACACCU ......(((((......)))))......((((((.((((-((.(((((((....)).((((......(((((............)))))......))))..))))).)))))).)))))) ( -34.20) >DroSec_CAF1 62238 120 + 1 UGCAUGUGCAUGCUUAAAUGCAAUAAAAGGGUGUCAAAUAGGAAUGCAGUGAAAACACAUAGUUGCAUACUCUCGAGAAGCAUUGAGUGAAAAUAUAUGGUUGCAUGCUUUUCAACACCU ......(((((......)))))......((((((......((.((((((((....))).....))))).))...(((((((((...(..(..(....)..)..)))))))))).)))))) ( -27.40) >DroSim_CAF1 66740 111 + 1 UGCAUGC---------AAUGCAAUUAAAGGGUGUCAAAUAGGAAUGCAGUGAAAACACAUAGUUGCAUACUCUCGAGAAGGAUUGAGUGAAAAUAUAUGGUUGCAUACUUUUCAACACCU (((((..---------.)))))......((((((.....(((.(((((((....)).((((......(((((............)))))......))))..))))).)))....)))))) ( -24.80) >DroEre_CAF1 67536 111 + 1 U----AUGC----UUGAAUGUAAUUAUGUGCAGUCAAAA-AGAAUGCUGUGAAAAUACAUAGUUGCUUACUCUCGAAGAGCAUCGAGUGAAAAUAUAUGUUUGCAUACUUUUCGACACCU .----..((----((((..(((((((((((...(((...-((....)).)))...)))))))))))...(((.....)))..))))))(((((..((((....)))).)))))....... ( -24.90) >DroYak_CAF1 65771 107 + 1 U----AUGCAUGCAUGAAUGCCAUUAUGGGCUGUCAAAA-AGAAUGCAG----AAUAAAUA--UGCACACUCUCGAAAAGCGUUGAGUGAAUAUAUAUGCUUGCAUACUUUU--ACACCU (----(((((.(((((...(((......)))........-....((((.----........--))))(((((.((.....))..)))))......))))).)))))).....--...... ( -25.90) >consensus UGCAUGUGCAUGCUUAAAUGCAAUUAAAGGGUGUCAAAA_AGAAUGCAGUGAAAACACAUAGUUGCAUACUCUCGAGAAGCAUUGAGUGAAAAUAUAUGGUUGCAUACUUUUCAACACCU ............................((((((......((.(((((((....)).((((......(((((............)))))......))))..))))).)).....)))))) (-12.78 = -14.74 + 1.96)

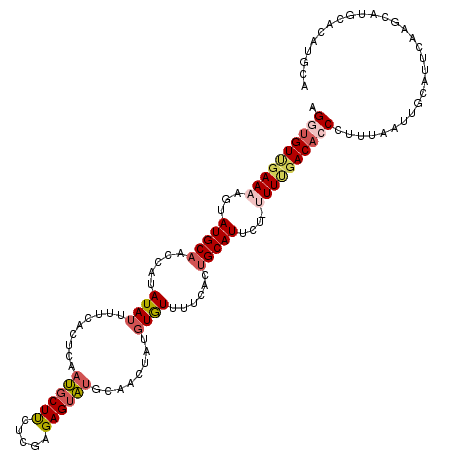
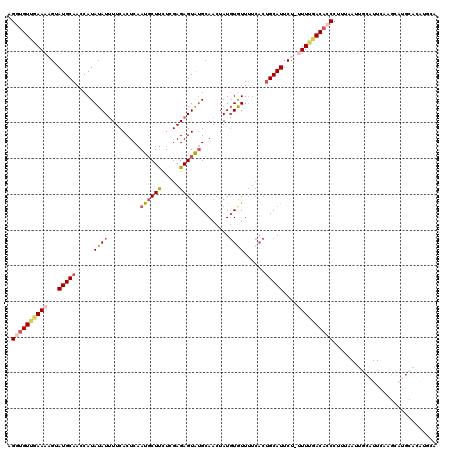
| Location | 24,239,590 – 24,239,709 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.56 |
| Mean single sequence MFE | -26.55 |
| Consensus MFE | -15.91 |
| Energy contribution | -17.15 |
| Covariance contribution | 1.24 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.60 |
| SVM decision value | 1.39 |
| SVM RNA-class probability | 0.949031 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24239590 119 - 27905053 AGGUGUUGAAAAGUAUGCAACCAUAUAUUUUCACUCAAUGCUUCUGGAGAGUAUGCAACUAUGUGUUUUCACUGCAUUCU-UUUCGACACCCUUUAAUUGCAUUUAAGCAUGCACAUGCA .((((((((((((.(((((....(((((((((.(...........)))))))))).......(((....)))))))).))-))))))))))................((((....)))). ( -34.50) >DroSec_CAF1 62238 120 - 1 AGGUGUUGAAAAGCAUGCAACCAUAUAUUUUCACUCAAUGCUUCUCGAGAGUAUGCAACUAUGUGUUUUCACUGCAUUCCUAUUUGACACCCUUUUAUUGCAUUUAAGCAUGCACAUGCA .(((((..((.((.(((((....(((((((((..............))))))))).......(((....))))))))..)).))..)))))................((((....)))). ( -26.94) >DroSim_CAF1 66740 111 - 1 AGGUGUUGAAAAGUAUGCAACCAUAUAUUUUCACUCAAUCCUUCUCGAGAGUAUGCAACUAUGUGUUUUCACUGCAUUCCUAUUUGACACCCUUUAAUUGCAUU---------GCAUGCA .(((((..((.((.(((((....(((((((((..............))))))))).......(((....))))))))..)).))..))))).......(((((.---------..))))) ( -25.24) >DroEre_CAF1 67536 111 - 1 AGGUGUCGAAAAGUAUGCAAACAUAUAUUUUCACUCGAUGCUCUUCGAGAGUAAGCAACUAUGUAUUUUCACAGCAUUCU-UUUUGACUGCACAUAAUUACAUUCAA----GCAU----A ..(((((((((((.((((...............(((((......)))))(((.....))).............)))).))-))))))).))................----....----. ( -23.10) >DroYak_CAF1 65771 107 - 1 AGGUGU--AAAAGUAUGCAAGCAUAUAUAUUCACUCAACGCUUUUCGAGAGUGUGCA--UAUUUAUU----CUGCAUUCU-UUUUGACAGCCCAUAAUGGCAUUCAUGCAUGCAU----A ......--....(((((((.(((...(((........((((((.....))))))...--....))).----.))).....-...(((..(((......)))..))))))))))..----. ( -22.96) >consensus AGGUGUUGAAAAGUAUGCAACCAUAUAUUUUCACUCAAUGCUUCUCGAGAGUAUGCAACUAUGUGUUUUCACUGCAUUCU_UUUUGACACCCUUUAAUUGCAUUCAAGCAUGCACAUGCA .((((((((((...(((((.....((((.........((((((.....))))))........))))......)))))....))))))))))............................. (-15.91 = -17.15 + 1.24)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:26:05 2006