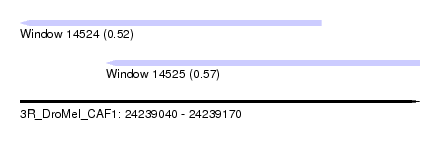
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 24,239,040 – 24,239,170 |
| Length | 130 |
| Max. P | 0.570971 |

| Location | 24,239,040 – 24,239,138 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 78.11 |
| Mean single sequence MFE | -16.61 |
| Consensus MFE | -9.49 |
| Energy contribution | -10.49 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.57 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.517768 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24239040 98 - 27905053 CAAGCGGCUUUUACAAGCAGUAAAUUUAAGUUGCUGCUGCUCCUAGCUACUUCCUGUUUUCUUUU---UUUUU-------CCA---CCCAUUUCGGCCCUUUUAUCCUCCG-- .(((..(((......((((((((.......))))))))......)))..))).............---.....-------...---.........................-- ( -16.20) >DroSec_CAF1 61688 98 - 1 CAAACGGCUUUUACAAGCAGUAAAUUUAAGUUGCUGCUGCUCCUUGCUACUUCCUGUUUUCUUUU---UUUUU-------CUA---CCGACUUCGGCCCUUUUAUCCUCCG-- .((((((........((((((((.......))))))))((.....))......))))))......---.....-------...---(((....)))...............-- ( -16.60) >DroSim_CAF1 66186 101 - 1 CAAGCGGCUUUUACAAGCAGUAAAUUUAAGUUGCUGCUGCUCCUUGCUACUUCCUGUUUUCUUUUUUUUUUUU-------CUA---CCGACUUCGGCCCUUUUAUCCUCCG-- .(((..((.......((((((((.......)))))))).......))..))).....................-------...---(((....)))...............-- ( -17.74) >DroEre_CAF1 66985 96 - 1 CAAGCGGCUUUUACAAGCAGUAAAUUUAAGUUGAUGCUGCUCCUUGCUACUUCCUGUUUUCUUU-----------CCCCCCCAC----CAUUUCGGCACUUUUAUCCUCCG-- .(((..((.......(((((((............)))))))....))..)))............-----------........(----(.....))...............-- ( -10.70) >DroYak_CAF1 65208 108 - 1 CAAGCGGCUUUUACAAGCAGUAAAUUUAAGUUGCUGCUGCUCCUUGCUACUUCCUGUUUUCCUUU---UUUUCCACCCCCCCACCUCCCAUUUUGGCCCUUUUAUCCUCCG-- .(((.((((......((((((((.......))))))))((.....))..................---..........................)))))))..........-- ( -16.40) >DroAna_CAF1 59745 93 - 1 CAAGCGGCUUUUACAAGCAGUAAAUUUAAGAUGAUUCUGCUUCG-GCUCCUGCUUUUGCUCCUAC---UCUUG----------------CCUGAGGAUCCUUCCGCUUCCCCC .((((((.......((((((........(((....)))((....-))..))))))....((((.(---.....----------------...))))).....))))))..... ( -22.00) >consensus CAAGCGGCUUUUACAAGCAGUAAAUUUAAGUUGCUGCUGCUCCUUGCUACUUCCUGUUUUCUUUU___UUUUU_______CCA___CCCACUUCGGCCCUUUUAUCCUCCG__ .(((.(((.......((((((((.......)))))))).......))).)))............................................................. ( -9.49 = -10.49 + 1.00)



| Location | 24,239,068 – 24,239,170 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 79.40 |
| Mean single sequence MFE | -21.66 |
| Consensus MFE | -12.74 |
| Energy contribution | -12.02 |
| Covariance contribution | -0.72 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.570971 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24239068 102 - 27905053 -----CCGCCUUUUGUUUUUAUUUACCCACACCGGGGCAAGCGGCUUUUACAAGCAGUAAAUUUAAGUUGCUGCUGCUCCUAGCUACUUCCUGUUUUCUUUU---UUUUU------ -----..(((((.(((............)))..)))))(((..(((......((((((((.......))))))))......)))..))).............---.....------ ( -21.50) >DroSim_CAF1 66214 105 - 1 -----CCGCCUUUUGUUUUUAUUUACCCACACCGGGGCAAGCGGCUUUUACAAGCAGUAAAUUUAAGUUGCUGCUGCUCCUUGCUACUUCCUGUUUUCUUUUUUUUUUUU------ -----....................(((.....)))(((((.(((.......((((((((.......))))))))))).)))))..........................------ ( -21.81) >DroEre_CAF1 67013 100 - 1 -----CCGCCUUUUGUUUUUAUUUACCCACACCGGGGCAAGCGGCUUUUACAAGCAGUAAAUUUAAGUUGAUGCUGCUCCUUGCUACUUCCUGUUUUCUUU-----------CCCC -----..(((.(((((.........(((.....)))))))).))).......(((((((............))))))).......................-----------.... ( -17.10) >DroWil_CAF1 21675 102 - 1 GGCUGGUCUCUUUUGUUUUUAUUUGCCCACACCGGAGCUGGCGGCUUUUACAAGCAGUAAAUUUAAGUUGAUGCUGCUUCCGCACAGUUUCCGCUUCCCC--------AU------ (((.....................))).....((((((((((((.......((((((((............)))))))))))).))).))))).......--------..------ ( -25.71) >DroYak_CAF1 65240 108 - 1 -----CCGCCUUUUGUUUUUAUUUACCCACACCGGGGCAAGCGGCUUUUACAAGCAGUAAAUUUAAGUUGCUGCUGCUCCUUGCUACUUCCUGUUUUCCUUU---UUUUCCACCCC -----....................(((.....)))(((((.(((.......((((((((.......))))))))))).)))))..................---........... ( -21.81) >DroAna_CAF1 59769 101 - 1 -----UCGUCUUUUGUUUUUAUUUACCCACACCGGAGCAAGCGGCUUUUACAAGCAGUAAAUUUAAGAUGAUUCUGCUUCG-GCUCCUGCUUUUGCUCCUAC---UCUUG------ -----............................(((((((((((.......((((((..((((......))))))))))..-....)))))..))))))...---.....------ ( -22.02) >consensus _____CCGCCUUUUGUUUUUAUUUACCCACACCGGGGCAAGCGGCUUUUACAAGCAGUAAAUUUAAGUUGAUGCUGCUCCUUGCUACUUCCUGUUUUCCUUU___UUUUU______ .................................(((((..((.((((....)))).)).......(((....)))))))).................................... (-12.74 = -12.02 + -0.72)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:26:00 2006