
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 24,228,285 – 24,228,386 |
| Length | 101 |
| Max. P | 0.630484 |

| Location | 24,228,285 – 24,228,386 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 80.62 |
| Mean single sequence MFE | -15.41 |
| Consensus MFE | -12.01 |
| Energy contribution | -11.73 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.24 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.630484 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24228285 101 - 27905053 UUUUCAAACAAAUUUACUGUCAUUAACUUAGACAGUUGAUGAAAUUUAUGUCAGACAUAUCAACAUCAUGGU-GACUCCAGACCAUCCGCCC----------ACCGCCCAAC ...............((((((.........))))))(((((.....((((.....))))....))))).(((-(.................)----------)))....... ( -15.53) >DroVir_CAF1 17500 94 - 1 UUUUCAAACAAAUUUACUGUCAUUAACUUAGACAGUUGAUGAAAUUUAUGUCAGACAUAUCAACGUCAUGAC-GUCCCUCAGCCUCCCCC-----------------CUCAA ................((((((((((((.....))))))))).....(((((((((........))).))))-))....)))........-----------------..... ( -17.40) >DroPse_CAF1 52430 102 - 1 UUUUCAAACAAAUUUACUGUCAUUAACUUAGACAGUUGAUGAAAUUUAUGUCAGACAUAUCAACAUCAUGGCCGGCUCCGUCUCCGUCUCCG----------ACUCCCACAC ...............((((((.........))))))(((((.....((((.....))))....)))))(((.(((...((....))...)))----------....)))... ( -15.70) >DroWil_CAF1 8684 101 - 1 UUUUCAAACAAAUUUACUGUCAUUAACUUAGACAGUUGAUGAAAUUUAUGUCAGACAUAUCAACAUCAUGGU-GACUCUUGCCCUACUCCAU----------ACACCCCCAC ...............((((((.........))))))(((((.....((((.....))))....))))).(((-(..................----------.))))..... ( -14.11) >DroAna_CAF1 49098 111 - 1 UUUUCAAACAAAUUUACUGUCAUUAACUUAGACAGUUGAUGAAAUUUAUGUCAGACAUAUCAACAUCAUGGU-GACUCCUGCCCCGCUACCCCUCUUCGUGCCCCGCAAAGC ...............((((((.........))))))(((((.....((((.....))))....))))).(..-..)...(((...((.((........))))...))).... ( -15.90) >DroPer_CAF1 52382 96 - 1 UUUUCAAACAAAUUUACUGUCAUUAACUUAGACAGUUGAUGAAAUUUAUGUCAGACAUAUCAACAUCAUGGCCGACUCCG------UCUCCG----------ACUCCCACAC ...............((((((.........))))))(((((.....((((.....))))....)))))(((..(((...)------)).)))----------.......... ( -13.80) >consensus UUUUCAAACAAAUUUACUGUCAUUAACUUAGACAGUUGAUGAAAUUUAUGUCAGACAUAUCAACAUCAUGGC_GACUCCGGCCCCGCCCCCC__________ACCCCCACAC ...(((.........((((((.........))))))(((((.....((((.....))))....))))))))......................................... (-12.01 = -11.73 + -0.28)

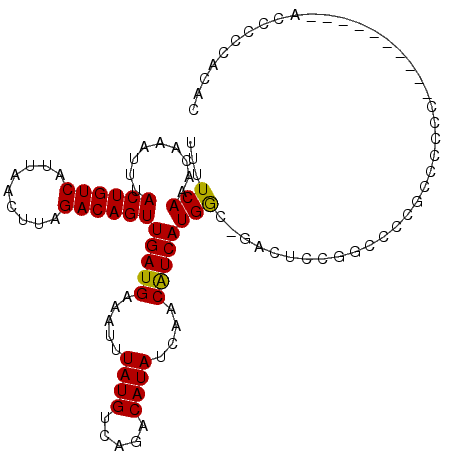

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:25:58 2006