
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 3,062,156 – 3,062,266 |
| Length | 110 |
| Max. P | 0.726339 |

| Location | 3,062,156 – 3,062,266 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.19 |
| Mean single sequence MFE | -30.55 |
| Consensus MFE | -17.00 |
| Energy contribution | -16.20 |
| Covariance contribution | -0.80 |
| Combinations/Pair | 1.27 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.726339 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3062156 110 + 27905053 -CGGCCG-UUCGAAU-UUGAG-CUCGCUUGCCAAUCAGCGGGCACAAAUUAUGGGCAAUUGUUUAAUUUGAAUAAGGCAGCGAUAGCUAUAGCAAUA------GCAAUAGCAACACCGAC -((((((-(((((((-(((.(-((((((........))))))).)))))).)))))..((((((.....))))))))).......(((((.((....------)).))))).....)).. ( -35.50) >DroSec_CAF1 49098 104 + 1 -CGGCCG-UUCGAAU-UUGAG-UCCGCUUGCCAAUCAGCGGGCACAAAUUAUGGGCAAUUGUUUAAUUUGAAUAAGGCAGCAAUAGCUAUAGCAA------------UAGCAGCACCGAC -((((((-(((((((-(((.(-((((((........))))))).)))))).)))))..((((((.....))))))))).((....(((((....)------------)))).))..)).. ( -32.60) >DroSim_CAF1 50293 116 + 1 -CGGCCG-UUCGAAU-UUGAG-CCCGCUUGCCAAUCAGCGGGCACAAAUUAUGGGCAAUUGUUUAAUUUGAAUAAGGCAGCAAUAGCUAUAGCAAUAGCAAUAGCAAUAGCAACACCGAC -((((((-(((((((-(((.(-((((((........))))))).)))))).)))))..((((((.....))))))))).((.((.(((((.((....)).))))).)).)).....)).. ( -40.80) >DroEre_CAF1 50459 98 + 1 -CGGCCG-UUCGAAU-UUGAG-UCCGAUUGCCAAUCAGCGGGCACAAAUUAUGGGCAAUUGUUUAAUUUGAAUAAGGCAGCCAUAG------------------CAAUAGCAUCAGCAAC -..((((-(((((((-(((.(-((((.(((.....)))))))).)))))).)))))..((((((.....))))))))).((....(------------------(....))....))... ( -25.90) >DroYak_CAF1 51104 98 + 1 -CGGCCG-UUCAAAU-UUGAG-CCCGAUUGCCAAUCAGCGGGCACAAAUUAUGGACAAUUGUUUAAUUUGAAUAAGGCAGAAAUAG------------------CAAUAACAACAGCAAC -..((((-(((((((-(((.(-((((.(((.....)))))))).)))))).)))))..((((((.....))))))))).......(------------------(..........))... ( -27.40) >DroPer_CAF1 49029 100 + 1 UGGGCCGAAUCGAAUUUUGAGACCCGGUUGCCAAUCAACGGACAUAAAUUAUGGGCAAUUGUUUAAUUUGUUUAAGGCAGCAAUAGC---AGCAA------------CAGCA-----AAC .((((((((......)))).).)))(((((((..((....))((((...)))))))))))......((((((....((.((....))---.))..------------.))))-----)). ( -21.10) >consensus _CGGCCG_UUCGAAU_UUGAG_CCCGCUUGCCAAUCAGCGGGCACAAAUUAUGGGCAAUUGUUUAAUUUGAAUAAGGCAGCAAUAGC___AGCAA_________CAAUAGCAACACCAAC ...(((..(((((((.(((.(.((((.(((.....)))))))).)))....(((((....))))))))))))...))).......................................... (-17.00 = -16.20 + -0.80)

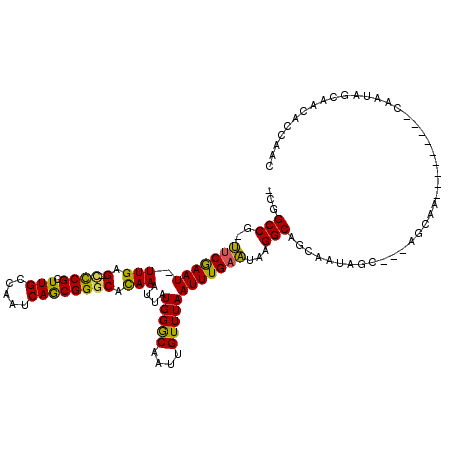
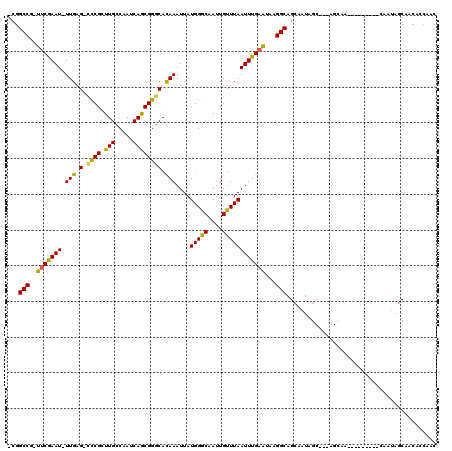
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:55:07 2006