
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 24,222,138 – 24,222,273 |
| Length | 135 |
| Max. P | 0.773868 |

| Location | 24,222,138 – 24,222,252 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.45 |
| Mean single sequence MFE | -31.62 |
| Consensus MFE | -13.18 |
| Energy contribution | -13.98 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.42 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.630885 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24222138 114 + 27905053 GCCCGUUGAAAUACGCUGAAGCGUUCGGCGCGAGCAUUUAAUUUAUGG--GCCAGCUGUUAAAUGUUGUACUUUGUGUGACAAAUAGCCUGCAAAGUUGCAUAUA----UGCACUUUGAG ((((((.((...((((....))))...((....))......)).))))--))((((((((...(((..(((...)))..)))))))).)))((((((.(((....----))))))))).. ( -35.30) >DroVir_CAF1 4979 110 + 1 GGU-GUAAUUUUCCGUAGAAAUGUUCAACGCCAGCGUUUAAUUUAUGA--GCCAGCUGUUAAGUGCUGCACUUUGUGUGACAAAUGGC-UACAAAGUUACAU--A----UGCAGCUUUGA (((-((...((((....))))......)))))((((((((.....)))--))..))).(((((.((((((...((((((((...((..-..))..)))))))--)----))))))))))) ( -27.20) >DroPse_CAF1 45271 99 + 1 GGCCCGUCCAUUGUGUUGAAACGU-------------UUAAUUUAUGG--GCCAGCUGUUAAAUGUUGCACUUUGUGUGACAAAUAGCCUGCAAAGUUGCAU--A----UGCACUUUGUA (((((((..(((((((....))).-------------.))))..))))--))).((((((...(((..(((...)))..))))))))).((((((((.((..--.----.)))))))))) ( -34.40) >DroWil_CAF1 1627 107 + 1 ACCCGUUC-AUUGUGUUGAAAUGUU------GAACAUUUAAUUUAUGGCGACCAGCUGUUAAAUGUUGCACUUUGUGUGACAAAUAGCUUCCAAAGUUGCAU--A----UGCACUUUGCA ...(((.(-((...((((((.((..------...))))))))..))))))...(((((((...(((..(((...)))..))))))))))..((((((.((..--.----.)))))))).. ( -26.30) >DroAna_CAF1 43112 118 + 1 GCCCGUCCAAAUGCGUUGAAACGUUCGGCGCGACCGUUUAAUUUAUGG--ACCAGCUGUUAAAUGUUGUACUUUGUGUGACAAAUAGCCUGCAAAGUUGCAUAUAUGUAUGCACGCUGCG ....(((((...(((((((.....)))))))((....))......)))--))..((((((...(((..(((...)))..)))))))))..(((..((.(((((....)))))))..))). ( -32.10) >DroPer_CAF1 45245 99 + 1 GGCCCGUCCAUUGUGUUGAAACGU-------------UUAAUUUAUGG--GCCAGCUGUUAAAUGUUGCACUUUGUGUGACAAAUAGCCUGCAAAGUUGCAU--A----UGCACUUUGUA (((((((..(((((((....))).-------------.))))..))))--))).((((((...(((..(((...)))..))))))))).((((((((.((..--.----.)))))))))) ( -34.40) >consensus GCCCGUUCAAUUGCGUUGAAACGUU______GA_C_UUUAAUUUAUGG__GCCAGCUGUUAAAUGUUGCACUUUGUGUGACAAAUAGCCUGCAAAGUUGCAU__A____UGCACUUUGCA ......................................................((((((...(((..(((...)))..)))))))))...((((((.(((........))))))))).. (-13.18 = -13.98 + 0.81)

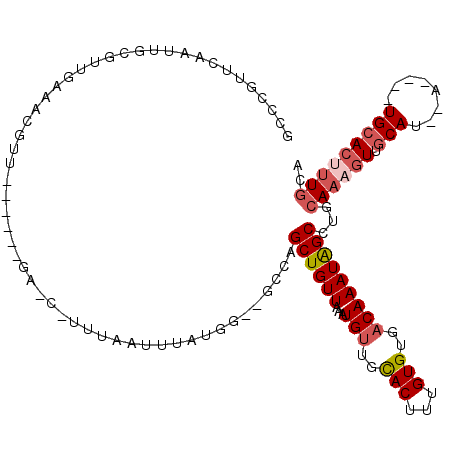

| Location | 24,222,178 – 24,222,273 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 81.04 |
| Mean single sequence MFE | -26.65 |
| Consensus MFE | -14.22 |
| Energy contribution | -15.05 |
| Covariance contribution | 0.83 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.55 |
| Structure conservation index | 0.53 |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.773868 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24222178 95 + 27905053 AUUUAUGG--GCCAGCUGUUAAAUGUUGUACUUUGUGUGACAAAUAGCCUGCAAAGUUGCAUAUA----UGCACUUUGAGAUUUUUCG-----GGCAC-----UGGUAAUU ........--(((((........(((..(((...)))..)))....((((.((((((.(((....----))))))))).((....)))-----))).)-----)))).... ( -26.20) >DroVir_CAF1 5018 102 + 1 AUUUAUGA--GCCAGCUGUUAAGUGCUGCACUUUGUGUGACAAAUGGC-UACAAAGUUACAU--A----UGCAGCUUUGAUUUUUAUAUGUUUUACAUAUAUAAGGCAAUU ........--(((....((((((.((((((...((((((((...((..-..))..)))))))--)----))))))))))))...((((((.....))))))...))).... ( -28.70) >DroEre_CAF1 49592 95 + 1 AUUUAUGG--GCCAGCUGUUAAAUGUUGGACUUUGUGUGACAAAUAGCCUGCAAAGUUGCAUAUA----UGCACUUUGAGAUUUUUCG-----GGCAC-----AGGUAAUU .....((.--(((.((((((...(((((.((...)).)))))))))))((.((((((.(((....----)))))))))))........-----))).)-----)....... ( -22.70) >DroWil_CAF1 1660 93 + 1 AUUUAUGGCGACCAGCUGUUAAAUGUUGCACUUUGUGUGACAAAUAGCUUCCAAAGUUGCAU--A----UGCACUUUGCAUUUUU--A-----UGCAA-----AGGUAAUU .......(((((.(((((((...(((..(((...)))..))))))))))......)))))..--.----(((.((((((((....--)-----)))))-----)))))... ( -29.50) >DroYak_CAF1 47469 95 + 1 AUUUAUGG--GCCAGCUGUUAAAUGUUGUACUUUGUGUGACAAAUAGCCUGCAAAGUUGCAUAUA----UGCACUUUGAGAUUUUUCG-----GGCAC-----AGGUAAUU .....((.--(((.((((((...(((..(((...)))..)))))))))((.((((((.(((....----)))))))))))........-----))).)-----)....... ( -24.80) >DroAna_CAF1 43152 99 + 1 AUUUAUGG--ACCAGCUGUUAAAUGUUGUACUUUGUGUGACAAAUAGCCUGCAAAGUUGCAUAUAUGUAUGCACGCUGCGAUUUGUUG-----GCCAA-----AAGUAAUU ((((.(((--.(((((.((((..(((..(((...)))..)))..)))).((((..((.(((((....)))))))..))))....))))-----)))).-----)))).... ( -28.00) >consensus AUUUAUGG__GCCAGCUGUUAAAUGUUGUACUUUGUGUGACAAAUAGCCUGCAAAGUUGCAUAUA____UGCACUUUGAGAUUUUUCG_____GGCAC_____AGGUAAUU ..........(((.((((((...((((((((...))))))))))))))...((((((.(((........)))))))))..........................))).... (-14.22 = -15.05 + 0.83)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:25:56 2006