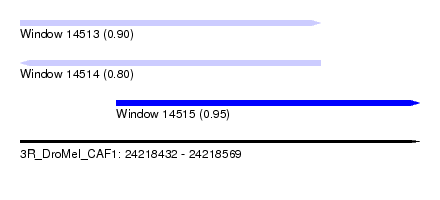
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 24,218,432 – 24,218,569 |
| Length | 137 |
| Max. P | 0.954007 |

| Location | 24,218,432 – 24,218,535 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 82.26 |
| Mean single sequence MFE | -27.13 |
| Consensus MFE | -18.75 |
| Energy contribution | -19.73 |
| Covariance contribution | 0.97 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.94 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.98 |
| SVM RNA-class probability | 0.895052 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
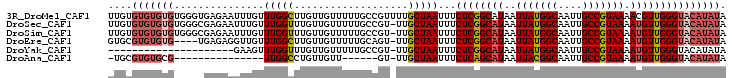
>3R_DroMel_CAF1 24218432 103 + 27905053 UUGUGUGUGUGUGGGUGAGAAUUUGUUUGGCUUGUUGUUUUUGCCGUUUUGCUAAUUUCUCGGCAUAAUUAUGGCAAUUGCCGUAAAACGUUGGGUACAUAUA .....(((((((((.((((((.((((.((((...........))))....)).)).)))))).)....(((((((....))))))).........)))))))) ( -26.80) >DroSec_CAF1 41083 102 + 1 UUGUGUGUGUGUGGGCGAGAAUUUGUUUGGUUUGUUGUUUUUGCCGU-UUGCUAAUUUCUCGGCAUAAUUAUGGCAAUUGCCGUAAAAUGUUGGGUACAUAUA .....(((((((((((((((((..............)))))))))..-..........((((((((..(((((((....))))))).)))))))))))))))) ( -30.84) >DroSim_CAF1 45072 102 + 1 UUGUGUGUGUGUGGGCGAGAAUUUGUUUGGUUUGUUGUUUUUGCCGU-UUGCUAAUUUCUCGGCAUAAUUAUGGCAAUUGCCGUAAAAUGUUGGGUACAUAUA .....(((((((((((((((((..............)))))))))..-..........((((((((..(((((((....))))))).)))))))))))))))) ( -30.84) >DroEre_CAF1 45854 98 + 1 GUGCGUGUGUG----UGAGAGGUUGUUUGGCUUGUUGUUUUUGCAGU-UUGCUAAUUUCUCGGCAUAAUUAUGGCAAUUGCCGUAAAAUGUUGGGUACAUAUA ((((.(((((.----((((((((....((((..(((((....)))))-..))))))))))))))))).(((((((....)))))))........))))..... ( -28.70) >DroYak_CAF1 43638 81 + 1 ---------------------GAAGUUUGGUUUGUUGUUUUUGCCGU-UUGCUAAUUUCUCGGCAUAAUUAUGGCAAUUGCCGUAAAAUGUUGGGUACAUAUA ---------------------..(((.((((..(.....)..)))).-..))).....((((((((..(((((((....))))))).))))))))........ ( -19.90) >DroAna_CAF1 38812 80 + 1 -UGCGUGUGCG---------------UUGGCCUGUUGUU------GU-UUGCUAAUUUCUCAGCAUAAUUACGGCAAUUGCCGUAAAAUGUUGGGUACAUAUA -...(((((((---------------(((((........------..-..))))))...(((((((..(((((((....))))))).)))))))))))))... ( -25.70) >consensus UUGUGUGUGUG_____GAGAAUUUGUUUGGCUUGUUGUUUUUGCCGU_UUGCUAAUUUCUCGGCAUAAUUAUGGCAAUUGCCGUAAAAUGUUGGGUACAUAUA ....(((((((...............(((((...................)))))...((((((((..(((((((....))))))).))))))))))))))). (-18.75 = -19.73 + 0.97)
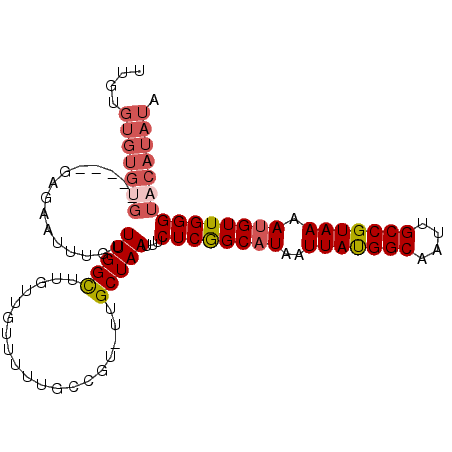
| Location | 24,218,432 – 24,218,535 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 82.26 |
| Mean single sequence MFE | -13.83 |
| Consensus MFE | -9.72 |
| Energy contribution | -9.72 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.798404 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24218432 103 - 27905053 UAUAUGUACCCAACGUUUUACGGCAAUUGCCAUAAUUAUGCCGAGAAAUUAGCAAAACGGCAAAAACAACAAGCCAAACAAAUUCUCACCCACACACACACAA ....(((.......(((((..(((....)))........((..(....)..)))))))(((...........)))..)))....................... ( -15.60) >DroSec_CAF1 41083 102 - 1 UAUAUGUACCCAACAUUUUACGGCAAUUGCCAUAAUUAUGCCGAGAAAUUAGCAA-ACGGCAAAAACAACAAACCAAACAAAUUCUCGCCCACACACACACAA ....(((..............(((....))).......(((((............-.)))))...)))................................... ( -11.82) >DroSim_CAF1 45072 102 - 1 UAUAUGUACCCAACAUUUUACGGCAAUUGCCAUAAUUAUGCCGAGAAAUUAGCAA-ACGGCAAAAACAACAAACCAAACAAAUUCUCGCCCACACACACACAA ....(((..............(((....))).......(((((............-.)))))...)))................................... ( -11.82) >DroEre_CAF1 45854 98 - 1 UAUAUGUACCCAACAUUUUACGGCAAUUGCCAUAAUUAUGCCGAGAAAUUAGCAA-ACUGCAAAAACAACAAGCCAAACAACCUCUCA----CACACACGCAC ....((((.............(((....))).......(((..(....)..))).-..))))..........................----........... ( -11.10) >DroYak_CAF1 43638 81 - 1 UAUAUGUACCCAACAUUUUACGGCAAUUGCCAUAAUUAUGCCGAGAAAUUAGCAA-ACGGCAAAAACAACAAACCAAACUUC--------------------- ....(((..............(((....))).......(((((............-.)))))...)))..............--------------------- ( -11.82) >DroAna_CAF1 38812 80 - 1 UAUAUGUACCCAACAUUUUACGGCAAUUGCCGUAAUUAUGCUGAGAAAUUAGCAA-AC------AACAACAGGCCAA---------------CGCACACGCA- .....((..((......(((((((....)))))))...((((((....)))))).-..------.......))...)---------------)((....)).- ( -20.80) >consensus UAUAUGUACCCAACAUUUUACGGCAAUUGCCAUAAUUAUGCCGAGAAAUUAGCAA_ACGGCAAAAACAACAAACCAAACAAAUUCUC_____CACACACACAA ....(((..............(((....))).......(((..(....)..)))...........)))................................... ( -9.72 = -9.72 + 0.00)

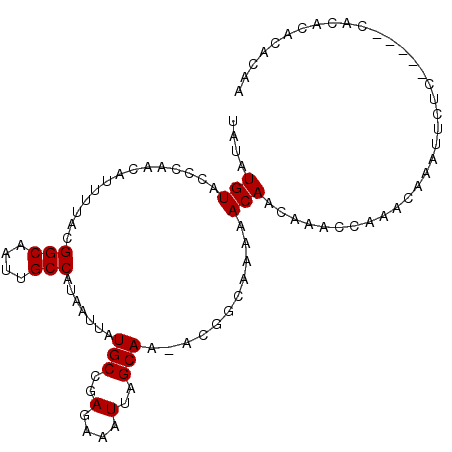
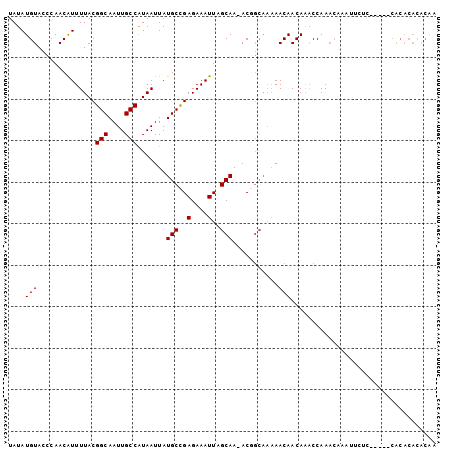
| Location | 24,218,465 – 24,218,569 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 94.19 |
| Mean single sequence MFE | -24.77 |
| Consensus MFE | -21.88 |
| Energy contribution | -21.93 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.44 |
| SVM RNA-class probability | 0.954007 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24218465 104 + 27905053 GUUGUUUUUGCCGUUUUGCUAAUUUCUCGGCAUAAUUAUGGCAAUUGCCGUAAAACGUUGGGUACAUAUAAAUUGGAAAUUAGUUAAUUUGCCGUUGCAUUGCU ..(((..(.((......((((((((((((((....(((((((....)))))))...)))))...((.......)))))))))))......)).)..)))..... ( -22.30) >DroSec_CAF1 41116 103 + 1 GUUGUUUUUGCCGU-UUGCUAAUUUCUCGGCAUAAUUAUGGCAAUUGCCGUAAAAUGUUGGGUACAUAUAAAUUGGAAAUUAGUUAAUUUGCCGUUGCAUUGCU ..(((..(.((.((-(.((((((((((((((((..(((((((....))))))).)))))))...((.......))))))))))).)))..)).)..)))..... ( -25.30) >DroSim_CAF1 45105 103 + 1 GUUGUUUUUGCCGU-UUGCUAAUUUCUCGGCAUAAUUAUGGCAAUUGCCGUAAAAUGUUGGGUACAUAUAAAUUGGAAAUUAGUUAAUUUACCGUUGCAUUGCU ..(((....((.((-..((((((((((((((((..(((((((....))))))).)))))))...((.......)))))))))))......)).)).)))..... ( -22.90) >DroEre_CAF1 45883 103 + 1 GUUGUUUUUGCAGU-UUGCUAAUUUCUCGGCAUAAUUAUGGCAAUUGCCGUAAAAUGUUGGGUACAUAUAAAUUGGAAAUUAGUUAAUUUUCUGUUGCAUCGCU (.(((....((((.-..((((((((((((((((..(((((((....))))))).)))))))...((.......))))))))))).......)))).))).)... ( -26.20) >DroYak_CAF1 43650 103 + 1 GUUGUUUUUGCCGU-UUGCUAAUUUCUCGGCAUAAUUAUGGCAAUUGCCGUAAAAUGUUGGGUACAUAUAAAUUGGAAAUUAGUUAAUUUGCCGUUGCAUCGCU (.(((..(.((.((-(.((((((((((((((((..(((((((....))))))).)))))))...((.......))))))))))).)))..)).)..))).)... ( -25.70) >DroAna_CAF1 38829 97 + 1 GUUGUU------GU-UUGCUAAUUUCUCAGCAUAAUUACGGCAAUUGCCGUAAAAUGUUGGGUACAUAUAAAUUGAAAAUUAGUUAAUUUGCAGUUGCAUUGCC ((..((------((-..((((((((((((((((..(((((((....))))))).))))))))..((.......)).))))))))......))))..))...... ( -26.20) >consensus GUUGUUUUUGCCGU_UUGCUAAUUUCUCGGCAUAAUUAUGGCAAUUGCCGUAAAAUGUUGGGUACAUAUAAAUUGGAAAUUAGUUAAUUUGCCGUUGCAUUGCU ..(((............((((((((((((((((..(((((((....))))))).)))))))...((.......)))))))))))............)))..... (-21.88 = -21.93 + 0.06)

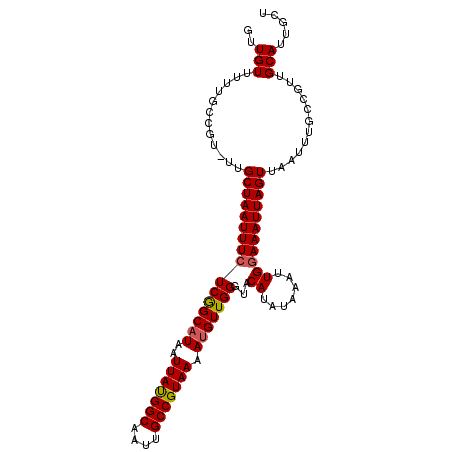
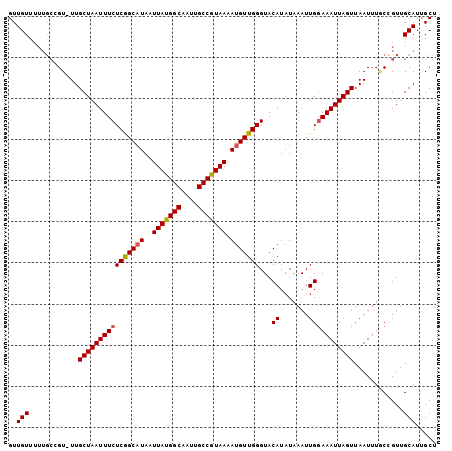
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:25:52 2006