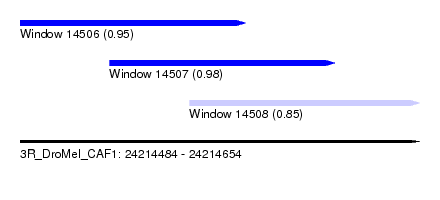
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 24,214,484 – 24,214,654 |
| Length | 170 |
| Max. P | 0.984673 |

| Location | 24,214,484 – 24,214,580 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 77.04 |
| Mean single sequence MFE | -18.81 |
| Consensus MFE | -4.28 |
| Energy contribution | -4.68 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.23 |
| SVM decision value | 1.43 |
| SVM RNA-class probability | 0.952932 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24214484 96 + 27905053 CGCCCGACUGCUGCAGUUUGAUGCACUCAAAAAA--AAACACAGCUUUGCUUCACAUAUU-UCGUCAUAGAAAAACUCCACA---AGCACGAA------AGUGCAUUA .((......((((..((((...............--)))).))))...))........((-((......)))).........---.((((...------.)))).... ( -16.46) >DroSec_CAF1 37092 98 + 1 CACCCGACUGCUGCAGUUUGCUGCACUCAAAAAAAAAAAAACAGAUAUGCUUCACAUAUU-UCGUCAUAGAAAAACUUCACA---AGCACGAA------AGUGCAUUA .....(((...(((((....))))).................(((((((.....))))))-).)))...(((....)))...---.((((...------.)))).... ( -22.10) >DroSim_CAF1 41045 94 + 1 CGCCCGACUGCUGCAGUUUGCUGCACUCAAAAA----AAACCAGAUAUGUUUCACAUAUU-UCGUCAUAGAAAAACUUCACA---AGCACGAA------AGUGCAUUA .....(((...(((((....)))))........----.....(((((((.....))))))-).)))...(((....)))...---.((((...------.)))).... ( -22.90) >DroEre_CAF1 41841 105 + 1 CGACCGAGUGCUGCAGUUUGCUGCACUACAAAAAA--AAACAAGAACUGCUUUACAUUUU-UCGCCAUGGAAAAAGUUCAAAAUAAUCACAAAACACUAAAAGCAUUU .....(((((((((((....))))...........--......(((((..(((.(((...-.....))).))).)))))......................))))))) ( -18.30) >DroYak_CAF1 39610 98 + 1 CGCCCGACUGCUGCAUUUUGCUGCACUAAAAAAAA--AAACAAAAUCUGCUUUACAUAUCUGUGUUGUAGAAAAAGUUUAAAAUAAUCACAAA--------CACAUUU .....((.(((.((.....)).)))..........--((((....(((((..((((....))))..)))))....)))).......)).....--------....... ( -14.30) >consensus CGCCCGACUGCUGCAGUUUGCUGCACUCAAAAAAA__AAACCAGAUAUGCUUCACAUAUU_UCGUCAUAGAAAAACUUCACA___AGCACGAA______AGUGCAUUA ...........(((((....)))))................................................................................... ( -4.28 = -4.68 + 0.40)

| Location | 24,214,522 – 24,214,618 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 70.55 |
| Mean single sequence MFE | -16.82 |
| Consensus MFE | -2.38 |
| Energy contribution | -2.26 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.55 |
| Structure conservation index | 0.14 |
| SVM decision value | 1.98 |
| SVM RNA-class probability | 0.984673 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24214522 96 + 27905053 ACAGCUUUGCUUCACAUAUU-UCGUCAUAGAAAAACUCCACA---AGCACGAA------AGUGCAUUAAAAGUGAUGGAAAAAG-CGAAAA-AACACAAAUCCAAAAA ...(((((...((((...((-((......)))).........---.((((...------.)))).......)))).....))))-).....-................ ( -21.10) >DroSec_CAF1 37132 97 + 1 ACAGAUAUGCUUCACAUAUU-UCGUCAUAGAAAAACUUCACA---AGCACGAA------AGUGCAUUAAAAGUGUUGGAAAAAC-CAAAUAGCACGCAAAUCCAAAAG ...(((.(((........((-((......)))).........---.((((...------.)))).......(((((((.....)-))....))))))).)))...... ( -20.60) >DroSim_CAF1 41081 97 + 1 CCAGAUAUGUUUCACAUAUU-UCGUCAUAGAAAAACUUCACA---AGCACGAA------AGUGCAUUAAAAGUGUUGGAAAAAA-CAAAUAGCACGCAAAUCCAAAAG ...(((.(((........((-((......)))).........---.((((...------.)))).......((((((.......-....))))))))).)))...... ( -17.70) >DroEre_CAF1 41879 107 + 1 CAAGAACUGCUUUACAUUUU-UCGCCAUGGAAAAAGUUCAAAAUAAUCACAAAACACUAAAAGCAUUUUAAGUGAUGGAAAAAACCAAAUGGCACACAAAUCCAAAAA ....................-..(((((..........................((((.(((....))).)))).(((......))).)))))............... ( -11.80) >DroYak_CAF1 39648 81 + 1 CAAAAUCUGCUUUACAUAUCUGUGUUGUAGAAAAAGUUUAAAAUAAUCACAAA--------CACAUUUUAGGUGAUGGAA-------------------AUCCAAAAA .....(((((..((((....))))..)))))..............(((((...--------..........)))))((..-------------------..))..... ( -12.92) >consensus CCAGAUAUGCUUCACAUAUU_UCGUCAUAGAAAAACUUCACA___AGCACGAA______AGUGCAUUAAAAGUGAUGGAAAAAA_CAAAUAGCACACAAAUCCAAAAA .............................................(((((.....................)))))(((.....................)))..... ( -2.38 = -2.26 + -0.12)

| Location | 24,214,556 – 24,214,654 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 71.29 |
| Mean single sequence MFE | -19.52 |
| Consensus MFE | -4.34 |
| Energy contribution | -4.82 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.22 |
| SVM decision value | 0.77 |
| SVM RNA-class probability | 0.847378 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24214556 98 + 27905053 CUCCACA---AGCACGAA------AGUGCAUUAAAAGUGAUGGAAAAAG-CGAAAA-AACACAAAUCCAAAAA-GGGCUUGGAAUUUAUAAUAACACUAU---GCAUUUGCUU .((((((---.((((...------.))))........)).))))..(((-((((..-....(((..((.....-))..))).....((((.......)))---)..))))))) ( -19.80) >DroSec_CAF1 37166 103 + 1 CUUCACA---AGCACGAA------AGUGCAUUAAAAGUGUUGGAAAAAC-CAAAUAGCACGCAAAUCCAAAAGGGGGGUAGGAAUUUAAAAUAACACUAUGUUGCAUUUGCUU .......---.((((...------.)))).......(((((((.....)-))....))))((((((.(((...((..((..............)).))...))).)))))).. ( -22.34) >DroSim_CAF1 41115 103 + 1 CUUCACA---AGCACGAA------AGUGCAUUAAAAGUGUUGGAAAAAA-CAAAUAGCACGCAAAUCCAAAAGUGGGGUAGGAAUUUAAAAUAACACCAUGUUGCAUUUGCUU .......---.((((...------.)))).......((((((.......-....))))))((((((.(((..((((.((..............)).)))).))).)))))).. ( -24.64) >DroEre_CAF1 41913 109 + 1 GUUCAAAAUAAUCACAAAACACUAAAAGCAUUUUAAGUGAUGGAAAAAACCAAAUGGCACACAAAUCCAAAAA-GGCGCUGGAAUUUAAAAUUACAGUCU---GCCUUUGCUU .........................((((.......(((.(((......))).....)))..........(((-(((((((.............))))..---)))))))))) ( -17.12) >DroYak_CAF1 39683 81 + 1 GUUUAAAAUAAUCACAAA--------CACAUUUUAGGUGAUGGAA-------------------AUCCAAAAA-GG-GUUGGAUUUUAAAAUUACAUUCU---GCCUUUGCUU ..............((((--------..((......(((((.(((-------------------((((((...-..-.)))))))))...)))))....)---)..))))... ( -13.70) >consensus CUUCACA___AGCACGAA______AGUGCAUUAAAAGUGAUGGAAAAAA_CAAAUAGCACACAAAUCCAAAAA_GGGGUUGGAAUUUAAAAUAACACUAU___GCAUUUGCUU ..........(((((.....................)))))........................(((((........)))))....................((....)).. ( -4.34 = -4.82 + 0.48)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:25:45 2006