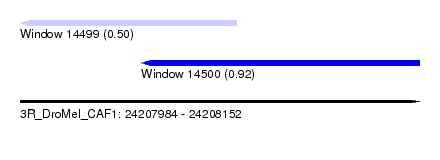
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 24,207,984 – 24,208,152 |
| Length | 168 |
| Max. P | 0.915432 |

| Location | 24,207,984 – 24,208,075 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.11 |
| Mean single sequence MFE | -26.23 |
| Consensus MFE | -10.95 |
| Energy contribution | -11.23 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.50 |
| Structure conservation index | 0.42 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
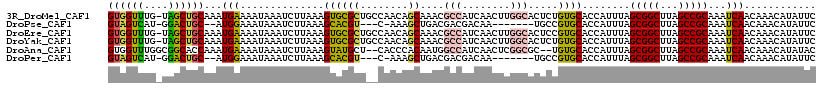
>3R_DroMel_CAF1 24207984 91 - 27905053 CACCAUUUAGCGGCUUAGCCGCAAAUCAACAAACAUAUUCG-UGAGCUCAAAG--------CCG-------------------CGGGCCAAGC-AAAAGCUAAACAAUAUUUUAGCAAAA ..((.....(((((((((((((.(((..........))).)-)).)))..)))--------)))-------------------).))......-....((((((......)))))).... ( -22.70) >DroPse_CAF1 31972 116 - 1 CACCAUUUAGCGGCUUAGCCGCAAAUCAACAAACAUAUUCG-UG-GCUCAACGGACUGCCGCCGGG-GGUAGUGCCGCUGCCGCUGCUGCUGC-AAAAGCUGAACAAUAUUUUAACAAAA .......(((((((..((((((.(((..........))).)-))-)))...(((((((((......-)))))).)))..)))))))..(((..-...))).................... ( -34.30) >DroEre_CAF1 35446 91 - 1 CACCAUUUAGCGGCUUAGCCGCAAAUCAACAAACAUAUUCG-UGAGCUCAAAG--------CCG-------------------CGGGCCAAGC-AAAAGCUAAACAAUAUUUUAGCAAAA ..((.....(((((((((((((.(((..........))).)-)).)))..)))--------)))-------------------).))......-....((((((......)))))).... ( -22.70) >DroYak_CAF1 32452 91 - 1 CACCAUUUAGCGGCUUAGCCGCAAAUCAACAAACAUAUUCG-UGAGCUCAAAG--------CCG-------------------CGGGCCAAGC-AAAAGCUAAACAAUAUUUUAGCAAAA ..((.....(((((((((((((.(((..........))).)-)).)))..)))--------)))-------------------).))......-....((((((......)))))).... ( -22.70) >DroAna_CAF1 28968 93 - 1 CACCAUUUAGCGGCUUAGCCGCAAAUCAACAAACAUAUACGCUUAGCUCAGCG--------CUG-------------------UGGGCCAAGCGAAAAGCUAAACAAUAUUUUAGGAAAA ..((.....(((((...))))).................(((((.(((((...--------...-------------------))))).)))))....................)).... ( -24.10) >DroPer_CAF1 31926 111 - 1 CACCAUUUAGCGGCUUAGCCGCAAAUCAACAAACAUAUUCG-UG-GCUCAACGGACUGCCGCCGGGGGGUAGUGCCG------CUGCUGCUGC-AAAAGCUGAACAAUAUUUUAACAAAA ........((((((..((((((.(((..........))).)-))-)))....((((((((.(....))))))).)))------)))))(((..-...))).................... ( -30.90) >consensus CACCAUUUAGCGGCUUAGCCGCAAAUCAACAAACAUAUUCG_UGAGCUCAAAG________CCG___________________CGGGCCAAGC_AAAAGCUAAACAAUAUUUUAGCAAAA .........(((((...)))))............................................................................((((((......)))))).... (-10.95 = -11.23 + 0.28)


| Location | 24,208,035 – 24,208,152 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 80.72 |
| Mean single sequence MFE | -30.47 |
| Consensus MFE | -19.24 |
| Energy contribution | -18.88 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.39 |
| Mean z-score | -2.62 |
| Structure conservation index | 0.63 |
| SVM decision value | 1.10 |
| SVM RNA-class probability | 0.915432 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
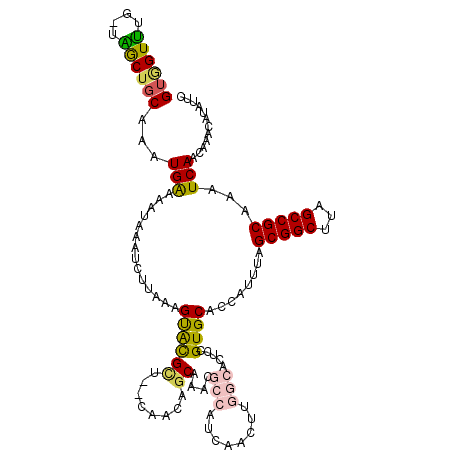
>3R_DroMel_CAF1 24208035 117 - 27905053 GUGGUUUG-UAGCUGCAAAUGAAAAUAAAUCUUAAAGUGCGCUGCCAACAGCAAACGCCAUCAACUUGGCACUCUGUGCACCAUUUAGCGGCUUAGCCGCAAAUCAACAAACAUAUUC ((((((..-.(((((((((((...............((((((((....))))....((((......)))).......))))))))).)))))).)))))).................. ( -36.06) >DroPse_CAF1 32048 104 - 1 GUAGUCAU-GGACUGC--AUGGAAAUAAAUCUUAAAGCACGU---C-AAAGCUGACGACGACAA-------UGCCGUGCACCAUUUAGCGGCUUAGCCGCAAAUCAACAAACAUAUUC ......((-((..(((--((((.............(((....---.-...)))...........-------..)))))))))))...(((((...))))).................. ( -24.87) >DroEre_CAF1 35497 117 - 1 GUGGUUUG-UAGCUGCAAAUGAAAAUAAAUCUUAAAGUGCGCUGCCAACAGCAAACGCCAUCAACUUGGCACUCCGUGCACCAUUUAGCGGCUUAGCCGCAAAUCAACAAACAUAUUC ((((((..-.(((((((((((...............((((((((....))))....((((......)))).......))))))))).)))))).)))))).................. ( -36.06) >DroYak_CAF1 32503 117 - 1 GUGGUUUG-UAGCUGCAAAUGAAAAUAAAUCUUAAAGUGCGCUGCCAACAGCAAACGCCAUCAACUUGGCACUCUGUGCACCAUUUAGCGGCUUAGCCGCAAAUCAACAAACAUAUUC ((((((..-.(((((((((((...............((((((((....))))....((((......)))).......))))))))).)))))).)))))).................. ( -36.06) >DroAna_CAF1 29021 114 - 1 GUGGUUUGGCGGCACCAAAUGAAAAUAAAUCUUAAAGUAUGCU--CACCCACAAUGGCCAUCAACUCGGCGC--UGUGCACCAUUUAGCGGCUUAGCCGCAAAUCAACAAACAUAUAC .(((((((.((((..............................--...........(((........)))((--(((..........)))))...)))))))))))............ ( -24.90) >DroPer_CAF1 31997 104 - 1 GUAGUCAU-GGACUGC--AUGGAAAUAAAUCUUAAAGCACGU---C-AAAGCUGACGACGACAA-------UGCCGUGCACCAUUUAGCGGCUUAGCCGCAAAUCAACAAACAUAUUC ......((-((..(((--((((.............(((....---.-...)))...........-------..)))))))))))...(((((...))))).................. ( -24.87) >consensus GUGGUUUG_UAGCUGCAAAUGAAAAUAAAUCUUAAAGUACGCU__CAACAGCAAACGCCAUCAACUUGGCACUCCGUGCACCAUUUAGCGGCUUAGCCGCAAAUCAACAAACAUAUUC ((((((....))))))...(((..............((((((........))....(((........))).....))))........(((((...)))))...)))............ (-19.24 = -18.88 + -0.36)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:25:39 2006