
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 24,205,722 – 24,205,817 |
| Length | 95 |
| Max. P | 0.786230 |

| Location | 24,205,722 – 24,205,817 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.22 |
| Mean single sequence MFE | -20.77 |
| Consensus MFE | -11.76 |
| Energy contribution | -11.76 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.653730 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24205722 95 + 27905053 A-AAAGGAAUAAGCGAAAAUUGAAUGUUUUUGUUGU--GUGGCCAGGCAAACUGGGCAA--------ACUUUAUGGCCACAAACACAAAUUACAACUACA------ACAUUU-------- .-.........(((((((((.....))))))))).(--(((((((.((.......))..--------......))))))))...................------......-------- ( -19.60) >DroPse_CAF1 29667 88 + 1 -------AGCAGGU-GAAAUUGAAUGUUUUUGUUGUGUGCGGCCAGGUA--CUGGGAAA--------ACUUUAUGGCCACAAACACAAAUUACAACUACA------ACAUUC-------- -------.......-......(((((((...((((((((.(((((....--..((....--------.))...))))).)).........))))))...)------))))))-------- ( -20.20) >DroGri_CAF1 2124 103 + 1 A-A----ACUGAAUAAAAAUUGAAUGUGUUUGUUGU--GCGGCCAGAUA--UAGGGCAA--------ACUUUAUGGCCACAAACACAAAUUACAACUACUAGCCCAACAACCAACAAACA .-.----.................(((((((((...--..(((((....--.(((....--------.)))..))))))))))))))................................. ( -19.10) >DroEre_CAF1 33077 95 + 1 A-AAAGGAAUAAGCGAAAAUUGAAUGUUUUUGUUGU--GUGGCCAGGCAAACUGGGCAA--------ACUUUAUGGCCACAAACACAAAUUACAACUACA------ACAUUU-------- .-.........(((((((((.....))))))))).(--(((((((.((.......))..--------......))))))))...................------......-------- ( -19.60) >DroYak_CAF1 30060 105 + 1 A-AAAGGAAUAAGCGAAAAUUGAAUGUUUUUGUUGUGUGUGGCCAGGCAAACUGGGCAAACUGGUAAACUUUAUGGCCACAAACACAAAUUACAACUACA------ACAUUU-------- .-...................(((((((...((((((((((((((.....((..(.....)..))........)))))))).........))))))...)------))))))-------- ( -25.92) >DroAna_CAF1 26655 90 + 1 AAA----AAUGAGUGAAAAUUGAAUGUUUUUGUUGU--GCGGCCAGGCA--GUGGGCAA--------ACUUUAUGGCCACAAACACAAAUUACAACUACA------ACAUUC-------- ...----..............(((((((...(((((--((......)).--((((.((.--------......)).))))...........)))))...)------))))))-------- ( -20.20) >consensus A_A____AAUAAGCGAAAAUUGAAUGUUUUUGUUGU__GCGGCCAGGCA__CUGGGCAA________ACUUUAUGGCCACAAACACAAAUUACAACUACA______ACAUUC________ ........................(((.(((((.((....(((((............................)))))....)))))))..))).......................... (-11.76 = -11.76 + -0.00)

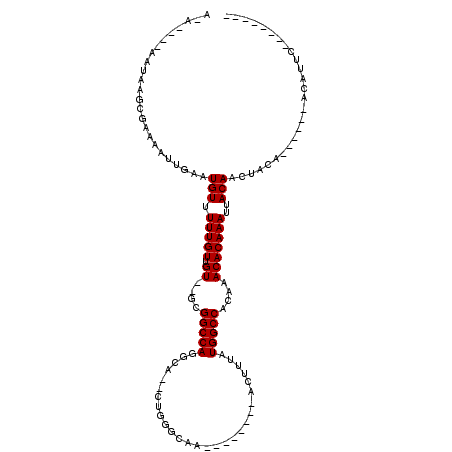
| Location | 24,205,722 – 24,205,817 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.22 |
| Mean single sequence MFE | -21.65 |
| Consensus MFE | -13.71 |
| Energy contribution | -13.46 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.786230 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24205722 95 - 27905053 --------AAAUGU------UGUAGUUGUAAUUUGUGUUUGUGGCCAUAAAGU--------UUGCCCAGUUUGCCUGGCCAC--ACAACAAAAACAUUCAAUUUUCGCUUAUUCCUUU-U --------....((------...(((((.......((((((((((((....((--------..((...))..)).)))))))--).))))........)))))...))..........-. ( -18.66) >DroPse_CAF1 29667 88 - 1 --------GAAUGU------UGUAGUUGUAAUUUGUGUUUGUGGCCAUAAAGU--------UUUCCCAG--UACCUGGCCGCACACAACAAAAACAUUCAAUUUC-ACCUGCU------- --------((((((------(....((((....(((((..(((((((......--------........--....)))))))))))))))).)))))))......-.......------- ( -23.87) >DroGri_CAF1 2124 103 - 1 UGUUUGUUGGUUGUUGGGCUAGUAGUUGUAAUUUGUGUUUGUGGCCAUAAAGU--------UUGCCCUA--UAUCUGGCCGC--ACAACAAACACAUUCAAUUUUUAUUCAGU----U-U (((((((((..(((..(((((((((..((((((((((........))))))..--------)))).)))--...))))))))--))))))))))...................----.-. ( -23.10) >DroEre_CAF1 33077 95 - 1 --------AAAUGU------UGUAGUUGUAAUUUGUGUUUGUGGCCAUAAAGU--------UUGCCCAGUUUGCCUGGCCAC--ACAACAAAAACAUUCAAUUUUCGCUUAUUCCUUU-U --------....((------...(((((.......((((((((((((....((--------..((...))..)).)))))))--).))))........)))))...))..........-. ( -18.66) >DroYak_CAF1 30060 105 - 1 --------AAAUGU------UGUAGUUGUAAUUUGUGUUUGUGGCCAUAAAGUUUACCAGUUUGCCCAGUUUGCCUGGCCACACACAACAAAAACAUUCAAUUUUCGCUUAUUCCUUU-U --------.(((((------(....((((....(((((..(((((((....((..((...........))..)).)))))))))))))))).))))))....................-. ( -20.90) >DroAna_CAF1 26655 90 - 1 --------GAAUGU------UGUAGUUGUAAUUUGUGUUUGUGGCCAUAAAGU--------UUGCCCAC--UGCCUGGCCGC--ACAACAAAAACAUUCAAUUUUCACUCAUU----UUU --------((((((------(...((((((((....))).(((((((...(((--------......))--)...)))))))--)))))...)))))))..............----... ( -24.70) >consensus ________AAAUGU______UGUAGUUGUAAUUUGUGUUUGUGGCCAUAAAGU________UUGCCCAG__UGCCUGGCCAC__ACAACAAAAACAUUCAAUUUUCACUUAUU____U_U .......................(((((...(((((((..(((((((............................)))))))..)).)))))......)))))................. (-13.71 = -13.46 + -0.25)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:25:36 2006