| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
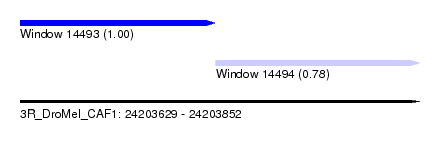
| Location | 24,203,629 – 24,203,852 |
| Length | 223 |
| Max. P | 0.995175 |

| Location | 24,203,629 – 24,203,738 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 87.36 |
| Mean single sequence MFE | -27.42 |
| Consensus MFE | -22.54 |
| Energy contribution | -22.06 |
| Covariance contribution | -0.48 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.69 |
| Structure conservation index | 0.82 |
| SVM decision value | 2.55 |
| SVM RNA-class probability | 0.995175 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24203629 109 + 27905053 UUCUCCGUUGGGUAUCACCCAAUAGUCCCUAGUUUUUAUCCCUCUUUAUUCGUUUUGUGUGUCAAAUGGCAUACAAUGGCUUACUCACAUGUGAGUCCGUUUUUACUGU ......((((((.....))))))......((((.......................(((((((....)))))))((((((((((......))))).)))))...)))). ( -26.80) >DroSec_CAF1 26375 103 + 1 ------GUUGGGUAUCAACCAAUAGUCCCUAGUUUUUACCCCUCUUUACUCGUUUUGUGUGUCAAAUGGCACACAAUGGCUUACUCACAUGUGAGUCCGUUUUUACUGU ------(..(((((..(((............)))..)))))..)...........((((((((....))))))))(((((((((......))))).))))......... ( -27.00) >DroSim_CAF1 30246 103 + 1 ------GUUGGGUAUCCACCAAUAGUCCCUAGUUUUUACCCCUCUUUACUCGUUUUGUGUGUCAAAUGGCACACAAUGGCUUACUCACAUGUGAGUCCGUUUUUACUGU ------(((((.......)))))......((((....((...............(((((((((....))))))))).(((((((......))))))).))....)))). ( -26.40) >DroEre_CAF1 30997 109 + 1 UUCUGCGGUAGGUUAUAUCCAAUGAUUCCUAGUUUUUGCCCCUCUUUAUUCGUUUUGUGUGUCAAAUGGCACACAAUGGCUUACUCACAUGUGAGUCCAUUUUUACUGU ....((((((((......))((((..............................(((((((((....))))))))).(((((((......)))))))))))..)))))) ( -27.30) >DroYak_CAF1 27909 109 + 1 UUCUCCGGUAGGAUUUAGCCAUGAGUUCCUAGUUUUUGCCCCUCUUUAUUCGUUUUGUGUGUCAAAUGGCACACAAUGGCUUACUCACAUGUGAGUCUGCUUUUACUGU .....((((((((...(((((((((...(........)...)))...........((((((((....)))))))))))))).(((((....)))))....)))))))). ( -29.60) >consensus UUCU_CGUUGGGUAUCAACCAAUAGUCCCUAGUUUUUACCCCUCUUUAUUCGUUUUGUGUGUCAAAUGGCACACAAUGGCUUACUCACAUGUGAGUCCGUUUUUACUGU ..........((......)).........((((....((...............(((((((((....))))))))).(((((((......))))))).))....)))). (-22.54 = -22.06 + -0.48)



| Location | 24,203,738 – 24,203,852 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.85 |
| Mean single sequence MFE | -30.03 |
| Consensus MFE | -19.86 |
| Energy contribution | -20.76 |
| Covariance contribution | 0.90 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.778563 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24203738 114 + 27905053 UAAAUGAAACACGUGCCAAGGAAGCAAUUUC-UGUGCAAUCUAUGACACAAGCGGUCCUUCGAUCUUU-GAAGCCUUC----GGCUCUUUGGUCACUUUGGCGCAUUUAAAACGCCACGG ............((((((((((.((......-((((((.....)).))))..(((..((((((...))-))))...))----)))))))))).)))..(((((.........)))))... ( -29.90) >DroPse_CAF1 27822 104 + 1 UAAAUGAAACACGUGCCAAGGAAGCAAUUUC-UGUGCAAUCUAUGACACAGACGGCCCUGC-----------GCCUUC----GGCUCUUUGGUCACUUUGGCACAUUUAAAACGCCACGA ............(((((((((........((-((((((.....)).)))))).((((..(.-----------(((...----))).)...)))).)))))))))................ ( -31.90) >DroGri_CAF1 429 106 + 1 UAAAUGAAACACGUUGGACGGAAGCAAUUUCGUGUGCAAUCUAUGGCGCAAACUGUGCC--------U-GAGGCCCUCUUGGCGCUCCUUGGUCGCUUUUACACAUUUUAAACGG----- ...........(((((.((((((....)))))).)((.(((...(((((.....)))))--------.-((((((.....))).)))...))).))..............)))).----- ( -27.00) >DroEre_CAF1 31106 114 + 1 UAAAUGAAACACGUGCCAAGGAAGCAAUUUC-UGUGCAAUCUAUGACACAAGCGGUCCUUCGAUCCUU-CAGGCCUCC----GGCUCCUUGGUCACUUUGGCGCAUUUAAAACGCCACGA ............((((((((((.((......-((((((.....)).))))..(((.(((..((....)-))))...))----)))))))))).)))..(((((.........)))))... ( -32.80) >DroAna_CAF1 24654 103 + 1 UAAAUGAAACACGUGCCAAGGAAGCAAUUUC-UGUGCAAUCUAUGACACAAGCGGUCC---GGUCCUUCGAAUCCUCC----GCCUCU---------UUGGCACAUUUAAAACGCCACGA ............((((((((((.((......-((((((.....)).))))((.(((.(---((....))).)))))..----)).)))---------)))))))................ ( -26.70) >DroPer_CAF1 27815 104 + 1 UAAAUGAAACACGUGCCAAGGAAGCAAUUUC-UGUGCAAUCUAUGACACAGACGGCCCUGC-----------GCCUUC----GGCUCUUUGGUCACUUUGGCACAUUUAAAACGCCACGA ............(((((((((........((-((((((.....)).)))))).((((..(.-----------(((...----))).)...)))).)))))))))................ ( -31.90) >consensus UAAAUGAAACACGUGCCAAGGAAGCAAUUUC_UGUGCAAUCUAUGACACAAACGGUCCU_C______U_GA_GCCUUC____GGCUCUUUGGUCACUUUGGCACAUUUAAAACGCCACGA ............(((((((((..(((........)))......((((.((((.((((.........................)))).)))))))))))))))))................ (-19.86 = -20.76 + 0.90)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:25:34 2006