| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 24,195,096 – 24,195,203 |
| Length | 107 |
| Max. P | 0.964408 |

| Location | 24,195,096 – 24,195,203 |
|---|---|
| Length | 107 |
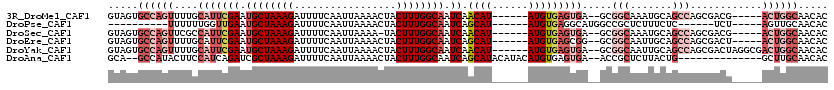
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.02 |
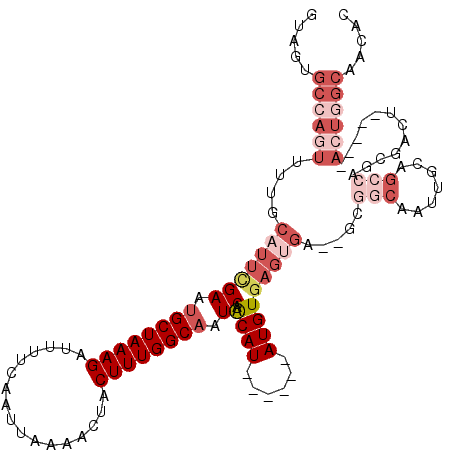
| Mean single sequence MFE | -29.79 |
| Consensus MFE | -15.42 |
| Energy contribution | -18.40 |
| Covariance contribution | 2.97 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.49 |
| Structure conservation index | 0.52 |
| SVM decision value | 1.57 |
| SVM RNA-class probability | 0.964408 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24195096 107 - 27905053 GUAGUGCCAGUUUUGCAUUCGAAUGCUAAAGAUUUUCAAUUAAAACUACUUUGGCAAUCAACAU------AUGUGAGUGA--GCGGCAAAUGCAGCCAGCGACG-----ACUGGCAACAC ((..((((((((((((((((((.((((((((.................)))))))).)).((..------..))))))..--..(((.......))).)))).)-----)))))))..)) ( -31.83) >DroPse_CAF1 19074 93 - 1 ----------UUUUUUGGUUGAAUGCUAAAGAUUUUCAAUUAAAACUACUUUGGCAAUCAGCAU------AUGUGAGGCAUGGCCGCUCUUUCUC------UCU-----AGUUGCAACAC ----------.......(((((.((((((((.................)))))))).)))))..------..(((.(((...)))((.((.....------...-----))..))..))) ( -19.83) >DroSec_CAF1 17933 106 - 1 GUAGUGCCAGUUCGCCAUUCGAAUGCUAAAGAUUUUCAAUUAAAA-UACUUUGGCAAUCAACAU------AUGUGAGUGA--GCGGCAAAUGCAGCCAGCGACG-----ACUGGCAACAC ((..((((((((((((((((((.(((((((((((((.....))))-).)))))))).)).((..------..))))))).--..(((.......))).))))..-----)))))))..)) ( -36.90) >DroEre_CAF1 21062 107 - 1 GUAGUGCCAGUUUUGCAUUCGAAUGCUAAAGAUUUUCAAUUAAAACUACUUUGGCAAUCAGCAU------AUGUGAGCGG--GCGGCAAUUGCAGCCAGCGACU-----ACUGGCAACAC ((..(((((((.((((....((.((((((((.................)))))))).)).((..------......)).(--((.((....)).))).))))..-----)))))))..)) ( -34.13) >DroYak_CAF1 19061 112 - 1 GUAGUGCCAGUUUUGCAUUCGAAUGCUAAAGAUUUUCAAUUAAAACUACUUUGGCAAUCAACAU------AUGUGAGUGA--GCGGCAAUUGCAGCCAGCGACUAGGCGACUGGCAACAC ((((((((.((....(((((((.((((((((.................)))))))).)).((..------..))))))).--))))).))))).(((((((......)).)))))..... ( -32.83) >DroAna_CAF1 16738 102 - 1 GCA--GCCAUACUUCCAUCAGAUCGCUAAAGAUUUUCAAUUAAAACUACUUUGGCAAUCAGCAUACAUACAUGUGAGUGA--ACCGCUCUUACUG--------------GCUUGCAACAC (((--((((...........(((.(((((((.................))))))).))).((((......))))(((((.--..)))))....))--------------)).)))..... ( -23.23) >consensus GUAGUGCCAGUUUUGCAUUCGAAUGCUAAAGAUUUUCAAUUAAAACUACUUUGGCAAUCAACAU______AUGUGAGUGA__GCGGCAAUUGCAGCCAGCGACU_____ACUGGCAACAC .....((((((....(((((((.((((((((.................)))))))).)).((((......))))))))).....(((.......)))............))))))..... (-15.42 = -18.40 + 2.97)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:25:29 2006