
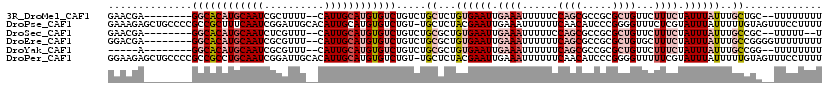
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 24,194,397 – 24,194,504 |
| Length | 107 |
| Max. P | 0.987824 |

| Location | 24,194,397 – 24,194,504 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 77.01 |
| Mean single sequence MFE | -31.69 |
| Consensus MFE | -14.88 |
| Energy contribution | -17.22 |
| Covariance contribution | 2.33 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.98 |
| Structure conservation index | 0.47 |
| SVM decision value | 2.09 |
| SVM RNA-class probability | 0.987824 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24194397 107 + 27905053 GAACGA--------GGCACAUGCAAUCGCUUUU--CAUUGCAUGUGUCUGUCUGCUCUGUGAAUUGAAAUUUUUCCAGCGCCGCGCUGUUCUUUCUAUUUAUUUGCUGC--UUUUUUUU .....(--------((((((((((((.......--.)))))))))))))((..((...((((((.((((......(((((...)))))...)))).))))))..)).))--........ ( -30.20) >DroPse_CAF1 18417 118 + 1 GAAAGAGCUGCCCCGCCGCUUUCAAUCGGAUUGCACAUUGCAUGUGUCUGU-UGCUCUACGAAUUGAAAUUUUUUCAACAUCCCGGGGUUUCUCGUAUUUAUUUUUGUAGUUUCCUUUU ....(((..((((((.((....(((.((((((((.....)))...))))))-)).....))..(((((.....))))).....))))))..)))......................... ( -24.60) >DroSec_CAF1 17261 105 + 1 GAACGA--------GGCACAUGCAAUCUCGUUU--CAUUGCAUGUGUCUGUCUGCGCUGUGAAUUGAAAUUUUUCCAGCGCCGCGCUGUUCUUUCUAUUUAUUUGCCGC--UUUUU--U ((((((--------((((((((((((.......--.)))))))))))))((..((((((.(((.........))))))))).))..)))))..................--.....--. ( -34.30) >DroEre_CAF1 20331 109 + 1 GGACGA--------GGCACAUGCAAUCGCGUUU--CAUUGCAUGUGUCUGUCUGCGCUGUGAAUUGAAAUUUUUUCAGCGCCGCGCUGUGCUUUCUAUUUAUUUGCCGGGGUUUUUUUU ((((.(--------((((((((((((.......--.)))))))))))))))))((((((.(((........))).)))))).((.(((.((.............))))).))....... ( -39.42) >DroYak_CAF1 18349 102 + 1 -----A--------GGCACAUGCAAUCGCGUUU--CAUUGCAUGUGUCUGUCUGCGCUGUGAAUUGAAAUUUUUUCAGCGCCGCGCUGUUCUUUCUAUUUAUUUGCCGG--UUUUUUUU -----(--------((((((((((((.......--.)))))))))))))..(((.((.((((((.((((......(((((...)))))...)))).))))))..)))))--........ ( -32.20) >DroPer_CAF1 18350 118 + 1 GGAAGAGCUGCCCCGCCGCCUGCAAUCGGAUUGCACAUUGCAUGUGUCUGU-UGCUCUACGAAUUGAAAUUUUUUCAACAUCCCGGGGUUUUUCGUAUUUAUUUUUGUAGUUUCCUUUU (((((((..((((((.((...((((.((((((((.....)))...))))))-)))....))..(((((.....))))).....))))))..)))....((((....)))).)))).... ( -29.40) >consensus GAACGA________GGCACAUGCAAUCGCGUUU__CAUUGCAUGUGUCUGUCUGCGCUGUGAAUUGAAAUUUUUUCAGCGCCGCGCUGUUCUUUCUAUUUAUUUGCCGA__UUUUUUUU ..............((((((((((((..........)))))))))))).....((...((((((.((((......((((.....))))...)))).))))))..))............. (-14.88 = -17.22 + 2.33)

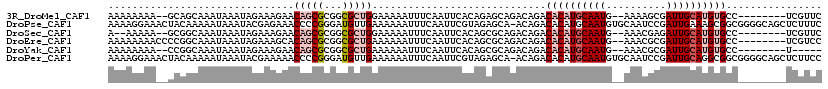
| Location | 24,194,397 – 24,194,504 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 77.01 |
| Mean single sequence MFE | -25.84 |
| Consensus MFE | -10.74 |
| Energy contribution | -12.02 |
| Covariance contribution | 1.28 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.42 |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.704090 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24194397 107 - 27905053 AAAAAAAA--GCAGCAAAUAAAUAGAAAGAACAGCGCGGCGCUGGAAAAAUUUCAAUUCACAGAGCAGACAGACACAUGCAAUG--AAAAGCGAUUGCAUGUGCC--------UCGUUC ........--...((.....(((.((((...(((((...)))))......)))).)))......)).(((((.((((((((((.--.......)))))))))).)--------).))). ( -24.10) >DroPse_CAF1 18417 118 - 1 AAAAGGAAACUACAAAAAUAAAUACGAGAAACCCCGGGAUGUUGAAAAAAUUUCAAUUCGUAGAGCA-ACAGACACAUGCAAUGUGCAAUCCGAUUGAAAGCGGCGGGGCAGCUCUUUC ...((....))..............(((...((((((..(((((.....................))-)))..)...(((.....)))..(((........))))))))...))).... ( -24.60) >DroSec_CAF1 17261 105 - 1 A--AAAAA--GCGGCAAAUAAAUAGAAAGAACAGCGCGGCGCUGGAAAAAUUUCAAUUCACAGCGCAGACAGACACAUGCAAUG--AAACGAGAUUGCAUGUGCC--------UCGUUC .--.....--(((.....................))).(((((((((.........))).)))))).(((((.((((((((((.--.......)))))))))).)--------).))). ( -29.20) >DroEre_CAF1 20331 109 - 1 AAAAAAAACCCCGGCAAAUAAAUAGAAAGCACAGCGCGGCGCUGAAAAAAUUUCAAUUCACAGCGCAGACAGACACAUGCAAUG--AAACGCGAUUGCAUGUGCC--------UCGUCC ...........((((..................)).))((((((................)))))).(((((.((((((((((.--.......)))))))))).)--------).))). ( -28.26) >DroYak_CAF1 18349 102 - 1 AAAAAAAA--CCGGCAAAUAAAUAGAAAGAACAGCGCGGCGCUGAAAAAAUUUCAAUUCACAGCGCAGACAGACACAUGCAAUG--AAACGCGAUUGCAUGUGCC--------U----- ........--..(((.......................((((((................))))))........(((((((((.--.......))))))))))))--------.----- ( -25.09) >DroPer_CAF1 18350 118 - 1 AAAAGGAAACUACAAAAAUAAAUACGAAAAACCCCGGGAUGUUGAAAAAAUUUCAAUUCGUAGAGCA-ACAGACACAUGCAAUGUGCAAUCCGAUUGCAGGCGGCGGGGCAGCUCUUCC ...((....))....................(((((....((((((.....))))))((((...(((-((.(((((((...)))))...)).).))))..))))))))).......... ( -23.80) >consensus AAAAAAAA__CCGGCAAAUAAAUAGAAAGAACAGCGCGGCGCUGAAAAAAUUUCAAUUCACAGAGCAGACAGACACAUGCAAUG__AAACGCGAUUGCAUGUGCC________UCGUUC ...............................(((((...))))).............................((((((((((..........))))))))))................ (-10.74 = -12.02 + 1.28)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:25:28 2006