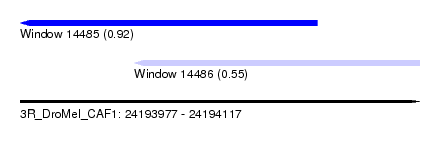
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 24,193,977 – 24,194,117 |
| Length | 140 |
| Max. P | 0.921857 |

| Location | 24,193,977 – 24,194,081 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.33 |
| Mean single sequence MFE | -21.32 |
| Consensus MFE | -11.52 |
| Energy contribution | -11.67 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.54 |
| SVM decision value | 1.14 |
| SVM RNA-class probability | 0.921857 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
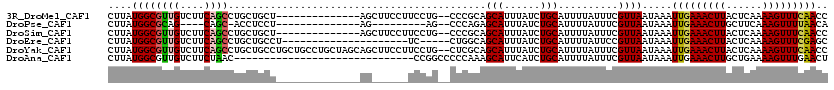
>3R_DroMel_CAF1 24193977 104 - 27905053 CUUAUGGCGUUGUCUUCAGCCUGCUGCU--------------AGCUUCCUUCCUG--CCCGCAGCAUUUAUCUGCAUUUUAUUUCGUUAAUAAAUUGAAACUUACUCAAAAGUUUCAACC ......((((((....)))).((((((.--------------.((.........)--)..)))))).......))..((((((.....))))))(((((((((......))))))))).. ( -23.90) >DroPse_CAF1 17929 90 - 1 CUUAUGGCGCAG----CAGC-ACCUCCU--------------AG---------AG--CCCAGAGCAUUUAUCUGCAUUUUAUUUCGUUAAUAAAUUGAAACUUGCUUCAAAGUUUUAACA .....((.((..----..))-.))...(--------------((---------((--..((((.......))))..))))).............(((((((((......))))))))).. ( -14.90) >DroSim_CAF1 20453 104 - 1 CUUAUGGCGUUGUCUUCAGCCUGCUGCU--------------AGCUUCCUUCCUG--CCCGCAGCAUUUAUCUGCAUUUUAUUUCGUUAAUAAAUUGAAACUUACUCAAAAGUUUCAACC ......((((((....)))).((((((.--------------.((.........)--)..)))))).......))..((((((.....))))))(((((((((......))))))))).. ( -23.90) >DroEre_CAF1 19909 94 - 1 CUUAUGGCGUUGUCUUCAGCCUGCUGCCU---------------------UC-----CUGGCAGCAUUUAUCUGCAUUUUAUUCCGUUAAUAAAUUGAAACUUACUCAAAAGUUUCGAGC ......((((((....)))).(((((((.---------------------..-----..))))))).......))...................(((((((((......))))))))).. ( -25.20) >DroYak_CAF1 17850 118 - 1 CUUAUGGCGUUGUCUUCAGCCUGCUGCCUGCUGCCUGCUAGCAGCUUCCUUCCUG--CUCGCAGCAUUUAUCUGCAUUUUAUUUCGUUAAUAAAUUGAAACUUACUCAAAAGUUUCAACC .....(((((((....))))..(((((..((.....))..))))).........)--)).((((.......))))..((((((.....))))))(((((((((......))))))))).. ( -28.90) >DroAna_CAF1 15525 90 - 1 CUUAUGGCGUUGUCUUCUAAC------------------------------CCGGCCCCCAAAGCAUUCAUCUGCAUUUUAUUUCGUUAAUAAAUUGAAACUUGCUGAAAAGUUUGAACU ....(((.((((.....))))------------------------------))).........(((......)))..((((((.....))))))((.((((((......)))))).)).. ( -11.10) >consensus CUUAUGGCGUUGUCUUCAGCCUGCUGCU______________AG______UCCUG__CCCGCAGCAUUUAUCUGCAUUUUAUUUCGUUAAUAAAUUGAAACUUACUCAAAAGUUUCAACC ....((((((((....))))...........................................(((......)))..........)))).....(((((((((......))))))))).. (-11.52 = -11.67 + 0.14)
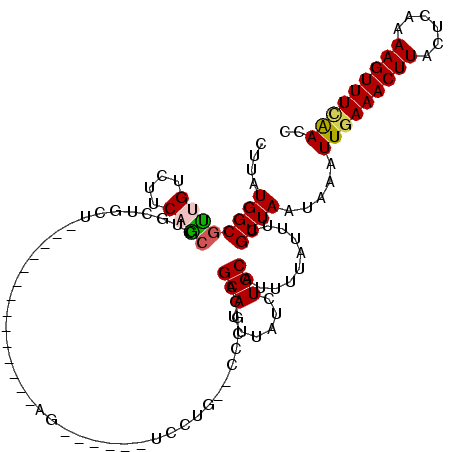

| Location | 24,194,017 – 24,194,117 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 80.65 |
| Mean single sequence MFE | -27.77 |
| Consensus MFE | -17.31 |
| Energy contribution | -18.14 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.550757 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
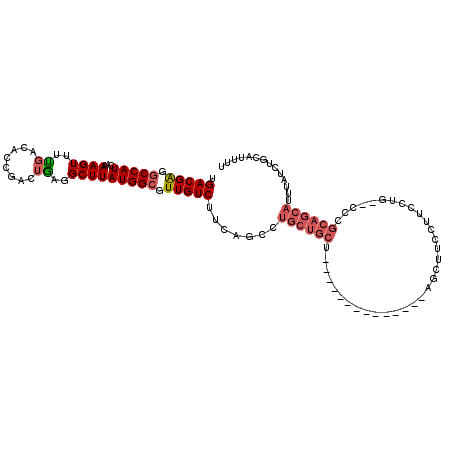
>3R_DroMel_CAF1 24194017 100 - 27905053 UGACGAGGCCAUCAAAAGUUUUGACACCGACUGAGGCUUAUGGCGUUGUCUUCAGCCUGCUGCU--------------AGCUUCCUUCCUG--CCCGCAGCAUUUAUCUGCAUUUU .(((((.(((((...((((((((........))))))))))))).)))))..(((..((((((.--------------.((.........)--)..)))))).....)))...... ( -28.90) >DroSec_CAF1 16879 100 - 1 UGACGAGGCCAUCAAAAGUUUUGACACCGACUGAGGCUUAUGGCGUUGUCUUCAGCCUGCUGCU--------------AGCUUCCUUCCUG--CCCGCAGCAUUUAUCUGCAUUUU .(((((.(((((...((((((((........))))))))))))).)))))..(((..((((((.--------------.((.........)--)..)))))).....)))...... ( -28.90) >DroSim_CAF1 20493 100 - 1 UGACGAGGCCAUCAAAAGUUUUGACACCGACUGAGGCUUAUGGCGUUGUCUUCAGCCUGCUGCU--------------AGCUUCCUUCCUG--CCCGCAGCAUUUAUCUGCAUUUU .(((((.(((((...((((((((........))))))))))))).)))))..(((..((((((.--------------.((.........)--)..)))))).....)))...... ( -28.90) >DroEre_CAF1 19949 90 - 1 UGACGAGGCCAUCAAAAGUUCUGACAGCGACUAAGGCUUAUGGCGUUGUCUUCAGCCUGCUGCCU---------------------UC-----CUGGCAGCAUUUAUCUGCAUUUU .(((((.(((((...((((..((........))..))))))))).)))))..(((..(((((((.---------------------..-----..))))))).....)))...... ( -29.90) >DroYak_CAF1 17890 114 - 1 UGACGAGGCCAUCAAAAGUUCUGACACCGACUAAGGCUUAUGGCGUUGUCUUCAGCCUGCUGCCUGCUGCCUGCUAGCAGCUUCCUUCCUG--CUCGCAGCAUUUAUCUGCAUUUU .(((((.(((((...((((..((........))..))))))))).)))))........(((((..((.....))..)))))..........--...((((.......))))..... ( -32.70) >DroAna_CAF1 15565 86 - 1 UGACGGGACCAUCAAAAGUUUCCACAUCGACUGCGGCUUAUGGCGUUGUCUUCUAAC------------------------------CCGGCCCCCAAAGCAUUCAUCUGCAUUUU ....(((.((.......((....))...(((.(((.(....).))).))).......------------------------------..))))).....(((......)))..... ( -17.30) >consensus UGACGAGGCCAUCAAAAGUUUUGACACCGACUGAGGCUUAUGGCGUUGUCUUCAGCCUGCUGCU______________AGCUUCCUUCCUG__CCCGCAGCAUUUAUCUGCAUUUU .(((((.(((((...((((..((........))..))))))))).))))).......((((((.................................)))))).............. (-17.31 = -18.14 + 0.84)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:25:26 2006