
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 24,193,220 – 24,193,325 |
| Length | 105 |
| Max. P | 0.710473 |

| Location | 24,193,220 – 24,193,325 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.40 |
| Mean single sequence MFE | -35.63 |
| Consensus MFE | -19.32 |
| Energy contribution | -20.52 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.98 |
| Structure conservation index | 0.54 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.592020 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
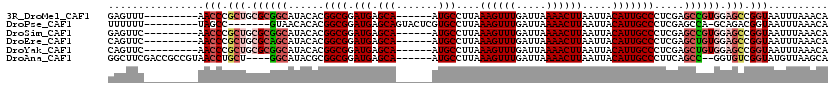
>3R_DroMel_CAF1 24193220 105 + 27905053 UGUUUAAAUUACCGGCUCCACGGCUCGAGGGCAAUGUAAUUAAGUUUUAAUCAAACUUUAAGGCAU------UGCUCAUCCGCCGUGUAUGCCGCGCAGCGGGUU---------AAACUC .((((((....(((((..((((((..(((((((((((....((((((.....))))))....))))------))))).)).))))))...)))((...)))).))---------)))).. ( -40.60) >DroPse_CAF1 17045 103 + 1 UGUUUAAAUUACCGUCUGC-UGGCUCGAGGGCAAUGUAAUUAAGUUUUAAUCAAACUUUAAGGCACGAGUACUGCUCAUCCGCCGUGUGUUAC-------GGCUA---------AAAAAA ........(((((((..((-((((..(((((((..(((.....(((((((.......))))))).....)))))))).)).)))).))...))-------)).))---------)..... ( -22.20) >DroSim_CAF1 19711 105 + 1 UGUUUAAAUUACCGGCUCCACGGCUCGAGGGCAAUGUAAUUAAGUUUUAAUCAAACUUUAAGGCAU------UGCUCAUCCGCCGUGUAUGCCGCGCAGCGGGUU---------GAACUC .((((((....(((((..((((((..(((((((((((....((((((.....))))))....))))------))))).)).))))))...)))((...)))).))---------)))).. ( -40.70) >DroEre_CAF1 19154 105 + 1 UGUUUAAAUUACCGGCUCCACAGCUCGAGGGCAAUGUAAUUAAGUUUUAAUCAAACUUUAAGGCAU------UGCUCAUCCGCCGUGUAUGCUGCGCAGCGGGUU---------GAACUG .((((((.....(((((....))).)).(((((((((....((((((.....))))))....))))------))))).(((((.((((.....)))).)))))))---------)))).. ( -35.40) >DroYak_CAF1 17045 105 + 1 UGUUUAAAUUACCGGCUCCACAGCUCGAGGGCAAUGUAAUUAAGUUUUAAUCAAACUUUAAGGCAU------UGCUCAUCCGCCGUGUAUGCCGCGCAGCGGGUU---------GAACUG .((((((.....(((((....))).)).(((((((((....((((((.....))))))....))))------))))).(((((.((((.....)))).)))))))---------)))).. ( -35.20) >DroAna_CAF1 14539 108 + 1 UGCUUAACAUACCGACACC--GGCUGAAGGGCAAUGUAAUUAAGUUUUAAUCAAACUUUAAGGCAU------UGCUCAUCCGCCGCGUAUGCC----AGCAGGUUACGGCGGUCGAAGCC .((((.....((((.(..(--(((.((.(((((((((....((((((.....))))))....))))------))))).)).))))((((.(((----....))))))))))))..)))). ( -39.70) >consensus UGUUUAAAUUACCGGCUCCACGGCUCGAGGGCAAUGUAAUUAAGUUUUAAUCAAACUUUAAGGCAU______UGCUCAUCCGCCGUGUAUGCCGCGCAGCGGGUU_________GAACUC ............((((..((((((....(((((((((....((((((.....))))))....))))......)))))....))))))...)))).......................... (-19.32 = -20.52 + 1.20)



| Location | 24,193,220 – 24,193,325 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.40 |
| Mean single sequence MFE | -32.43 |
| Consensus MFE | -22.94 |
| Energy contribution | -24.27 |
| Covariance contribution | 1.32 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.710473 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24193220 105 - 27905053 GAGUUU---------AACCCGCUGCGCGGCAUACACGGCGGAUGAGCA------AUGCCUUAAAGUUUGAUUAAAACUUAAUUACAUUGCCCUCGAGCCGUGGAGCCGGUAAUUUAAACA ..((((---------((.....(((.((((...((((((.((.(.(((------(((.....((((((.....)))))).....))))))).))..))))))..)))))))..)))))). ( -36.50) >DroPse_CAF1 17045 103 - 1 UUUUUU---------UAGCC-------GUAACACACGGCGGAUGAGCAGUACUCGUGCCUUAAAGUUUGAUUAAAACUUAAUUACAUUGCCCUCGAGCCA-GCAGACGGUAAUUUAAACA ......---------..(((-------((.....)))))(((((((.....))))).))...((((((.....))))))..(((.((((((.((......-...)).)))))).)))... ( -24.40) >DroSim_CAF1 19711 105 - 1 GAGUUC---------AACCCGCUGCGCGGCAUACACGGCGGAUGAGCA------AUGCCUUAAAGUUUGAUUAAAACUUAAUUACAUUGCCCUCGAGCCGUGGAGCCGGUAAUUUAAACA ......---------.......(((.((((...((((((.((.(.(((------(((.....((((((.....)))))).....))))))).))..))))))..)))))))......... ( -33.90) >DroEre_CAF1 19154 105 - 1 CAGUUC---------AACCCGCUGCGCAGCAUACACGGCGGAUGAGCA------AUGCCUUAAAGUUUGAUUAAAACUUAAUUACAUUGCCCUCGAGCUGUGGAGCCGGUAAUUUAAACA ......---------.(((.(((.((((((.....(((.((....(((------(((.....((((((.....)))))).....))))))))))).)))))).))).))).......... ( -30.70) >DroYak_CAF1 17045 105 - 1 CAGUUC---------AACCCGCUGCGCGGCAUACACGGCGGAUGAGCA------AUGCCUUAAAGUUUGAUUAAAACUUAAUUACAUUGCCCUCGAGCUGUGGAGCCGGUAAUUUAAACA ......---------.......(((.((((...((((((.((.(.(((------(((.....((((((.....)))))).....))))))).))..))))))..)))))))......... ( -31.70) >DroAna_CAF1 14539 108 - 1 GGCUUCGACCGCCGUAACCUGCU----GGCAUACGCGGCGGAUGAGCA------AUGCCUUAAAGUUUGAUUAAAACUUAAUUACAUUGCCCUUCAGCC--GGUGUCGGUAUGUUAAGCA (((.......))).((((.((((----(((((...((((.((.(.(((------(((.....((((((.....)))))).....)))))).).)).)))--)))))))))).)))).... ( -37.40) >consensus GAGUUC_________AACCCGCUGCGCGGCAUACACGGCGGAUGAGCA______AUGCCUUAAAGUUUGAUUAAAACUUAAUUACAUUGCCCUCGAGCCGUGGAGCCGGUAAUUUAAACA ................(((.(((.((((((......((((.(((.(((.......)))....((((((.....)))))).....))))))).....)))))).))).))).......... (-22.94 = -24.27 + 1.32)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:25:25 2006