| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 24,188,571 – 24,188,683 |
| Length | 112 |
| Max. P | 0.559273 |

| Location | 24,188,571 – 24,188,683 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.29 |
| Mean single sequence MFE | -30.46 |
| Consensus MFE | -16.39 |
| Energy contribution | -17.23 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.54 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.559273 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24188571 112 - 27905053 UGGCUGCGAAAUAUGCAGCC---AGCAGACUGACAAAUCGCU--UACCUUUAAAUCCGGUAGC-CUGCAAACUGCUUUAA-GGGCUCC-AGAGCCGUGAAGCUGCACAACAUUCAUAUUU ((((((((.....)))))))---)((((...((....))(((--.(((.........))))))-)))).....((((((.-.((((..-..)))).)))))).................. ( -34.50) >DroSec_CAF1 11478 112 - 1 UGGCUGCGAAAUAUGCAGCC---AGCGGACUGACAAAUCGCU--CACCUUUAAAUCCAGUAGC-CUGCAAGCUGCUUUAA-GGGCUCU-ACAGCCAUGAAGCUGCACAACAUUCAUAUUU ((((((((.....)))))))---)((((((((..........--............))))...-))))..((.((((((.-.((((..-..)))).)))))).))............... ( -32.45) >DroSim_CAF1 15108 112 - 1 UGGCUGCGAAAUAUGCAGCC---AGCGGACUGACAAAUCGCU--CACCUUUAAAUCCAGCAGC-CUGCAAACUGCUUUAA-GGGCUCC-AGAGCCAUGAAGCUGCACAACAUUCAUAUUU ((((((((.....)))))))---)((((.((.....((.(((--(.(((((((....(((((.-.......)))))))))-)))....-.)))).))..))))))............... ( -33.50) >DroEre_CAF1 14428 114 - 1 UGGCUGCGAAAUAUGCAGCC---AGCGGACUGACAAAUCGCUCUCACCUUUAAAUCCGGUAGC-CAAAAGACAGCUUUAA-GGGCUCC-AGUGCCUGGAAGCUGCACAACAUUCAUAUUU ((((((((.....)))))))---)...((.((.......(((...(((.........))))))-.....(.(((((((..-((((...-...)))).))))))))....)).))...... ( -32.30) >DroYak_CAF1 12288 114 - 1 UUGCUGCGAAAUAUGCAGCC---AGCGGACUGACAAAUCGCU--CACCAUUAAAUCCAGUAGCCCAAAAAACGGCUUUAACUGGCUCG-AGUGCCUGGAAGCUGCACAACAUUCAUAUUU ..((((((.....)))))).---.((((.((..((...((((--(..........(((((((((........))))...)))))...)-))))..))..))))))............... ( -30.12) >DroAna_CAF1 9964 104 - 1 UGCCUGUGAAAUAUUCAGGCAACAGCAAACUGACAAAUAACU--CACCUUUAAGCCCAU------------GUUCUUUAA-AGGGACUAAGUAACAGGACGGUGAA-AUCAUAAAUACUU ((((((.........)))))).(((....))).........(--((((....(((((..------------.........-.))).))..((......))))))).-............. ( -19.90) >consensus UGGCUGCGAAAUAUGCAGCC___AGCGGACUGACAAAUCGCU__CACCUUUAAAUCCAGUAGC_CUGCAAACUGCUUUAA_GGGCUCC_AGAGCCAGGAAGCUGCACAACAUUCAUAUUU .(((((((.....)))))))....(((.((((........................)))).............(((((....(((((...)))))..))))))))............... (-16.39 = -17.23 + 0.84)

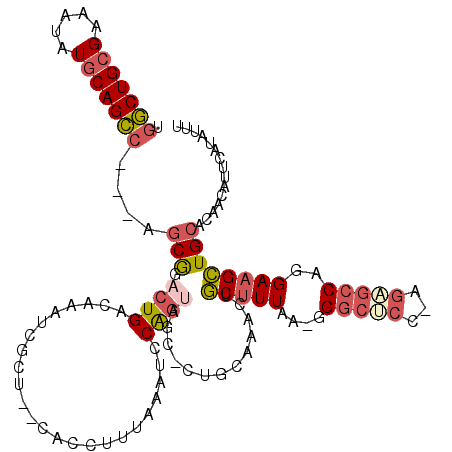

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:25:19 2006