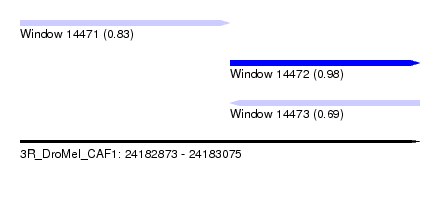
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 24,182,873 – 24,183,075 |
| Length | 202 |
| Max. P | 0.978164 |

| Location | 24,182,873 – 24,182,979 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 81.03 |
| Mean single sequence MFE | -19.02 |
| Consensus MFE | -15.10 |
| Energy contribution | -15.10 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.826627 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24182873 106 + 27905053 UCGCCCCUGGUCCACCGA----------AAAAUGUGUAUGCAAAAAUGCAAUAAGCGAGCACAGCUUAAAUAAAAUGCAUUUCAAUGAUUGCAUUUUGGAUUAAAGCAACAAACAU ..((...((((((.....----------......(((((......))))).(((((.......)))))...((((((((..........))))))))))))))..))......... ( -21.90) >DroPse_CAF1 5254 101 + 1 UCGGA----------CAA--AAGGUUUCCAAAUUUGUAUGCAAAAAUGCAAUAAGCGAGUGCAGCUUAAAUAAAAUGCAUUUCAAUGAUUGCAUUUUGGAUUAAAACAACAAC--- .....----------...--......(((((.((((....))))((((((((....(((((((............))))))).....))))))))))))).............--- ( -19.60) >DroSec_CAF1 5888 105 + 1 UCGACCCUGUCC-CCCGA----------AAAAUGUGUAUGCAAAAAUGCAAUAAGCGAGCACAGCUUAAAUAAAAUGCAUUUCAAUGAUUGCAUUUUGGAUUAAAGCAACAAACAU ............-.....----------....(((((.(((.............))).)))))((((...(((((((((..........))))))))).....))))......... ( -19.02) >DroEre_CAF1 8794 92 + 1 --------------CCAA----------AAAAUGUGUAUGCAAAAAUGCAAUAAGCGAGCGCAGCUUAAAUAAAAUGCAUUUCAAUGAUUGCAUUUUGGAUUAAACCAACAAACAU --------------....----------.....((((.((((....)))).(((((.......)))))..(((((((((..........)))))))))..............)))) ( -16.00) >DroYak_CAF1 6587 105 + 1 UCGUUAGGGGC-CACCAA----------AAAAUGUAUAUGCAAAAAUGCAAUAAGCGAGCGCAGCUUAAAUAAAAUGCAUUUCAAUGAUUGCAUUUUGGAUUAAACCAACAAACAU ..(((.((...-..((((----------(((((((((.((((....)))).(((((.......)))))......))))))))..(((....))))))))......)))))...... ( -19.50) >DroAna_CAF1 4507 104 + 1 ------------CACCACACAAGCCUUUCAAAUUUGUAUGCAAAAAUGCAAUAAGCGAGUGCAGCUUAAAUAAAAUGCAUUUCAAUGAUUGCAUUUUGGAUUAAAACAACAAAAAU ------------..((......((....((....))...)).((((((((((....(((((((............))))))).....))))))))))))................. ( -18.10) >consensus UCG__________ACCAA__________AAAAUGUGUAUGCAAAAAUGCAAUAAGCGAGCGCAGCUUAAAUAAAAUGCAUUUCAAUGAUUGCAUUUUGGAUUAAAACAACAAACAU ......................................((((....)))).(((((.......)))))..(((((((((..........))))))))).................. (-15.10 = -15.10 + -0.00)


| Location | 24,182,979 – 24,183,075 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 78.49 |
| Mean single sequence MFE | -19.47 |
| Consensus MFE | -10.66 |
| Energy contribution | -11.62 |
| Covariance contribution | 0.96 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.50 |
| Structure conservation index | 0.55 |
| SVM decision value | 1.81 |
| SVM RNA-class probability | 0.978164 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24182979 96 + 27905053 -UCGACAGGGCAC-AACAAUAACAUCGAGAACAGUCGACAAUUCUUUGUGCAAAACC--------UGCAUACAUACUC-GUACUAUAUACAGUACCCCAUGCCCAUU -......(((((.-............(((.......((....))..((((((.....--------))))))....)))-(((((......)))))....)))))... ( -20.60) >DroSec_CAF1 5993 96 + 1 -UCGACAGGGCAC-AACAAUAACAUCGAGAACAGUCGACAGUUCUUUGUGCAAAACC--------UGCAUACAUACCC-GUACUAUACACAGUAUCCCAUGCCCAUU -......(((((.-............((((((........))))))((((((.....--------)))))).......-(((((......)))))....)))))... ( -20.90) >DroSim_CAF1 5019 96 + 1 -UCGACAGGGCAC-AACAAUAACAUCGAGAACAGUCGACAGUUCUUUGUGCAAAACC--------UGCAUACAUACCC-GUACUAUACACAGUAUCCCAUGCCCAUU -......(((((.-............((((((........))))))((((((.....--------)))))).......-(((((......)))))....)))))... ( -20.90) >DroEre_CAF1 8886 93 + 1 UUCGACUGGGCAC-AACAAUAACAUCCACAGCAGUCGACAUCUCUUUGUGCAAUACCUAC--UCGUACAUACAUACCC-GCACUGUAUA----------UACCCAUU ......(((((((-((........((.((....)).)).......)))))).........--..(((.(((((.....-....))))).----------)))))).. ( -13.06) >DroYak_CAF1 6692 100 + 1 UUCGACUGGGCACCAACAAUAACAGCCAGAGACAUCGACAUCUCUUUGUGCAAAUCGUACAAACAUACAUACAUACCCCUUACCAUAUAGAG-------UGCCCAUU ......((((((((.............(((((........)))))((((((.....))))))...........................).)-------)))))).. ( -21.90) >consensus _UCGACAGGGCAC_AACAAUAACAUCGAGAACAGUCGACAGUUCUUUGUGCAAAACC________UGCAUACAUACCC_GUACUAUAUACAGUA_CCCAUGCCCAUU .......(((((..............((((((........))))))((((((.............))))))............................)))))... (-10.66 = -11.62 + 0.96)



| Location | 24,182,979 – 24,183,075 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 78.49 |
| Mean single sequence MFE | -28.52 |
| Consensus MFE | -15.54 |
| Energy contribution | -15.26 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.54 |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.689624 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24182979 96 - 27905053 AAUGGGCAUGGGGUACUGUAUAUAGUAC-GAGUAUGUAUGCA--------GGUUUUGCACAAAGAAUUGUCGACUGUUCUCGAUGUUAUUGUU-GUGCCCUGUCGA- ....((((.((....(((((((((....-.....))))))))--------).....((((((.....((((((......))))))......))-))))))))))..- ( -27.30) >DroSec_CAF1 5993 96 - 1 AAUGGGCAUGGGAUACUGUGUAUAGUAC-GGGUAUGUAUGCA--------GGUUUUGCACAAAGAACUGUCGACUGUUCUCGAUGUUAUUGUU-GUGCCCUGUCGA- ....((((.((....(((((((((....-.....))))))))--------).....((((((..(((.(((((......))))))))....))-))))))))))..- ( -27.80) >DroSim_CAF1 5019 96 - 1 AAUGGGCAUGGGAUACUGUGUAUAGUAC-GGGUAUGUAUGCA--------GGUUUUGCACAAAGAACUGUCGACUGUUCUCGAUGUUAUUGUU-GUGCCCUGUCGA- ....((((.((....(((((((((....-.....))))))))--------).....((((((..(((.(((((......))))))))....))-))))))))))..- ( -27.80) >DroEre_CAF1 8886 93 - 1 AAUGGGUA----------UAUACAGUGC-GGGUAUGUAUGUACGA--GUAGGUAUUGCACAAAGAGAUGUCGACUGCUGUGGAUGUUAUUGUU-GUGCCCAGUCGAA ..((((((----------((.(((((((-((.(((.(((......--))).))))))))).....((((((.((....)).))))))..))))-)))))))...... ( -29.10) >DroYak_CAF1 6692 100 - 1 AAUGGGCA-------CUCUAUAUGGUAAGGGGUAUGUAUGUAUGUUUGUACGAUUUGCACAAAGAGAUGUCGAUGUCUCUGGCUGUUAUUGUUGGUGCCCAGUCGAA ..((((((-------((....(..((((.(((((.((.(((((....))))))).)))....((((((......))))))..)).))))..).))))))))...... ( -30.60) >consensus AAUGGGCAUGGG_UACUGUAUAUAGUAC_GGGUAUGUAUGCA________GGUUUUGCACAAAGAAAUGUCGACUGUUCUCGAUGUUAUUGUU_GUGCCCUGUCGA_ ...((((((........(((((((.(((...))))))))))...............(((...(((((........)))))...)))........))))))....... (-15.54 = -15.26 + -0.28)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:25:14 2006