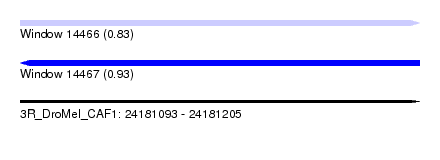
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 24,181,093 – 24,181,205 |
| Length | 112 |
| Max. P | 0.934834 |

| Location | 24,181,093 – 24,181,205 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 72.98 |
| Mean single sequence MFE | -23.69 |
| Consensus MFE | -2.66 |
| Energy contribution | -2.34 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.11 |
| SVM decision value | 0.72 |
| SVM RNA-class probability | 0.833283 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24181093 112 + 27905053 CGA-GCAUUCCGGCCAUUUUUUAUUGCCAAAUGUCCGUUCAACUCGACCGAUUGAAUCACCGCCACAG--GACGCUU-UCUCAUUUUUG-CUUCUCUUUGCGUCGGCUUAAAUUUUU ((.-.((((..(((.((.....)).))).))))..))(((((.((....))))))).....(((....--(((((..-...........-.........)))))))).......... ( -20.85) >DroSec_CAF1 4140 99 + 1 CGA-GCGUUCCGGCCAUUUUUUAUUGCCAAAUGGCCAUUCAGCUCGAC------------CACCACAG--GACGCUC-UCUCAUU-UUG-CUUUCCUUUGCGACGGCUUAAAUUUUU .((-((((((.((((((((.........))))))))..((.....)).------------.......)--)))))))-.......-..(-((.((......)).))).......... ( -25.30) >DroSim_CAF1 3159 99 + 1 CGA-GCGUUCCGGCCAUUUUUUAUUGCCAAAUGGCCAUUCAGCUCGAC------------CACCACAG--GACGCUC-UCUCAUU-UUG-CUUUCCUUUGCGACGGCUUAAAUUUUU .((-((((((.((((((((.........))))))))..((.....)).------------.......)--)))))))-.......-..(-((.((......)).))).......... ( -25.30) >DroEre_CAF1 7111 102 + 1 CGA-GCAUUCCGGCCAUUUUUUAUGGCCAAAUGACCGUUCGACUCGAC------------CGAUUGAGCAGCCGCUUUUCUAAUU-UUG-CUCCUUUUUGCGGUGGCUUACAUUUUC ...-.......((((((.....))))))(((((...((((((.((...------------.))))))))(((((((.........-..(-(........)))))))))..))))).. ( -27.60) >DroPer_CAF1 3326 94 + 1 CGAGGCAUUCCGUUCAUUUUUUAUUGUAA-CGCUCCAUUGAAUCCGG----------------GG-CG--GACGCUU-UCUAAUU-UUGGGUUUCUUUUUUGGUUCCUUACAUUUU- .(((((..((((((((........)).))-)(((((.........))----------------))-))--)).))))-)......-..(((..((......))..)))........- ( -19.40) >consensus CGA_GCAUUCCGGCCAUUUUUUAUUGCCAAAUGGCCAUUCAACUCGAC____________CACCACAG__GACGCUU_UCUCAUU_UUG_CUUUCCUUUGCGACGGCUUAAAUUUUU .........................(((...((......))...............................(((........................)))..))).......... ( -2.66 = -2.34 + -0.32)


| Location | 24,181,093 – 24,181,205 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 72.98 |
| Mean single sequence MFE | -26.48 |
| Consensus MFE | -9.06 |
| Energy contribution | -9.94 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.34 |
| SVM decision value | 1.24 |
| SVM RNA-class probability | 0.934834 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
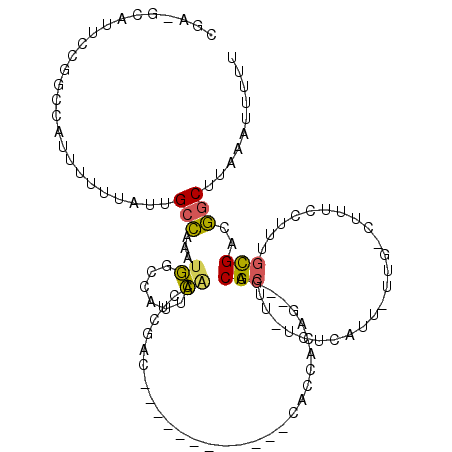
>3R_DroMel_CAF1 24181093 112 - 27905053 AAAAAUUUAAGCCGACGCAAAGAGAAG-CAAAAAUGAGA-AAGCGUC--CUGUGGCGGUGAUUCAAUCGGUCGAGUUGAACGGACAUUUGGCAAUAAAAAAUGGCCGGAAUGC-UCG ..........(((((.((........)-).....((((.-.(.((((--....)))).)..)))).)))))(((((....(((.(((((.........))))).)))....))-))) ( -27.70) >DroSec_CAF1 4140 99 - 1 AAAAAUUUAAGCCGUCGCAAAGGAAAG-CAA-AAUGAGA-GAGCGUC--CUGUGGUG------------GUCGAGCUGAAUGGCCAUUUGGCAAUAAAAAAUGGCCGGAACGC-UCG ..........(((..((((..(((..(-(..-.......-..)).))--)))))..)------------))(((((....(((((((((.........)))))))))....))-))) ( -32.20) >DroSim_CAF1 3159 99 - 1 AAAAAUUUAAGCCGUCGCAAAGGAAAG-CAA-AAUGAGA-GAGCGUC--CUGUGGUG------------GUCGAGCUGAAUGGCCAUUUGGCAAUAAAAAAUGGCCGGAACGC-UCG ..........(((..((((..(((..(-(..-.......-..)).))--)))))..)------------))(((((....(((((((((.........)))))))))....))-))) ( -32.20) >DroEre_CAF1 7111 102 - 1 GAAAAUGUAAGCCACCGCAAAAAGGAG-CAA-AAUUAGAAAAGCGGCUGCUCAAUCG------------GUCGAGUCGAACGGUCAUUUGGCCAUAAAAAAUGGCCGGAAUGC-UCG ..........((....))......(((-((.-.....((....((((((......))------------))))..)).........(((((((((.....))))))))).)))-)). ( -28.00) >DroPer_CAF1 3326 94 - 1 -AAAAUGUAAGGAACCAAAAAAGAAACCCAA-AAUUAGA-AAGCGUC--CG-CC----------------CCGGAUUCAAUGGAGCG-UUACAAUAAAAAAUGAACGGAAUGCCUCG -....(((((.(..(((.....(.....)..-.......-..(.(((--((-..----------------.))))).)..)))..).-))))).............((....))... ( -12.30) >consensus AAAAAUUUAAGCCGCCGCAAAGGAAAG_CAA_AAUGAGA_AAGCGUC__CUGUGGCG____________GUCGAGUUGAAUGGACAUUUGGCAAUAAAAAAUGGCCGGAAUGC_UCG .......................................................................(((((....(((((((((.........)))))))))....)).))) ( -9.06 = -9.94 + 0.88)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:25:09 2006