
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 24,180,372 – 24,180,467 |
| Length | 95 |
| Max. P | 0.843395 |

| Location | 24,180,372 – 24,180,467 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 73.80 |
| Mean single sequence MFE | -26.90 |
| Consensus MFE | -18.80 |
| Energy contribution | -18.97 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.21 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.843395 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
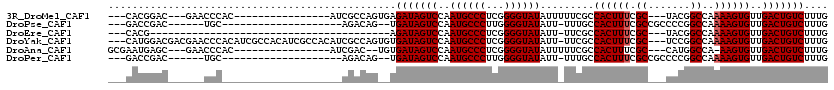
>3R_DroMel_CAF1 24180372 95 - 27905053 ---CACGGAC---GAACCCAC----------------AUCGCCAGUGAGAUAGUCCAAUGCCCUCGGGGUAUAUUUUUCGCCACUUUCGC---UACGGCCAAAAGUGUUGACUGUCUUUG ---(((((.(---((......----------------.))))).)))((((((((..((((((...)))))).........((((((.((---....))..))))))..))))))))... ( -30.10) >DroPse_CAF1 2588 88 - 1 ---GACCGAC------UGC--------------------AGACAG--UGAUAGUCCAAUGCCCUUGGGGUAUAUU-UUUGCCACUUUCGCCGCCCCGGCCAAAAGUGUUGACUGUCUUUG ---.....((------((.--------------------...)))--)(((((((..((((((...))))))...-.....((((((.((((...))))..))))))..))))))).... ( -26.90) >DroEre_CAF1 6434 73 - 1 ---CACG----------------------------------------AGAUAGUCCAAUGCCCUCGGGGUAUAUU-UUCGCCACUUUCGC---UACGGCCAAAAGUGUUGACUGUCUUUG ---...(----------------------------------------((((((((..((((((...))))))...-.....((((((.((---....))..))))))..))))))))).. ( -25.20) >DroYak_CAF1 3997 113 - 1 ---CAUGGACGACGAACCCACAUCGCCACAUCGCCACAUCGCCAGUGUGAUAGUCCAAUGCCCUCGGGGUAUAUU-UUCGCCACUUUCGC---UCCGGCCAAAAGUGUUGACUGUCUUUG ---..(((((..(((.......))).(((((.((......))..)))))...)))))((((((...))))))...-.(((.((((((.((---....))..)))))).)))......... ( -26.80) >DroAna_CAF1 2170 95 - 1 GCGAAUGAGC---GAACCCAC----------------AUCGAC--UGUGAUAGUCCAAUGCCCUCGGGGUAUAUUUUUCGCCACUUUCGC---CAUGGCCA-AAGUGUUGACUGUCUUUG ((......))---........----------------......--...(((((((..((((((...)))))).........((((((.((---....)).)-)))))..))))))).... ( -25.50) >DroPer_CAF1 2575 88 - 1 ---GACCGAC------UGC--------------------AGACAG--UGAUAGUCCAAUGCCCUUGGGGUAUAUU-UUUGCCACUUUCGCCGCCCCGGCCAAAAGUGUUGACUGUCUUUG ---.....((------((.--------------------...)))--)(((((((..((((((...))))))...-.....((((((.((((...))))..))))))..))))))).... ( -26.90) >consensus ___CACGGAC______CCC____________________CGACAG__UGAUAGUCCAAUGCCCUCGGGGUAUAUU_UUCGCCACUUUCGC___CACGGCCAAAAGUGUUGACUGUCUUUG ................................................(((((((..((((((...)))))).........((((((.((.......))..))))))..))))))).... (-18.80 = -18.97 + 0.17)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:25:08 2006