
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 24,171,805 – 24,171,935 |
| Length | 130 |
| Max. P | 0.986736 |

| Location | 24,171,805 – 24,171,895 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 90 |
| Reading direction | forward |
| Mean pairwise identity | 86.93 |
| Mean single sequence MFE | -29.96 |
| Consensus MFE | -21.52 |
| Energy contribution | -21.04 |
| Covariance contribution | -0.48 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.94 |
| Structure conservation index | 0.72 |
| SVM decision value | 1.65 |
| SVM RNA-class probability | 0.969959 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24171805 90 + 27905053 UUAGGCUGUGGGUUGUUGUUGUGGCUGCCGUUGCAAGCGGCAGGUGCAGCAUUCCACAGAAAGAAAUUUUGUAAGAAAUAUCUAUAUCUA .(((((((((((....(((((((.(((((((.....))))))).))))))).)))))))..(((.(((((....))))).)))...)))) ( -35.60) >DroSec_CAF1 12417 89 + 1 UUGGGCUGUGGGUUGUUGUUA-GGCUGCCGUUGCAAGUUGCAGGUGCAGCAUUCCACAGAAAGAAAAUAUGUCAGAAAAUUCCAAAUCUU (((((((((((..((((((..-(.((((..(.....)..)))).)))))))..))))))...((.......)).......)))))..... ( -24.30) >DroSim_CAF1 12357 90 + 1 UUGGGCUGUGGGUUGUUGUUGCGGCUGCCGUUGCAAGUUGCAGGUGCAGCAUUCCACAGAAAGAAAAUAUGUCAGAAAAUUCUAAAUCUA (((((((((((..(((((((((((((((....)).)))))))...))))))..))))))...((.......)).......)))))..... ( -27.60) >DroEre_CAF1 5686 87 + 1 UUGGGCUGUGGGUUGUUGU---GGCUGUCGUUGCAAGUUGCAGGUGCAGCAUUCCACAGAAAACAAAUGUGGCAUAAAAUGCAAUAUCUA (((..((((((..((((((---(.((((..(.....)..)))).)))))))..))))))....)))((((.((((...)))).))))... ( -28.10) >DroYak_CAF1 5942 87 + 1 UUGGGCUGUGGGUUGUUGU---GGCUGUCGUUGCAAGCUGCAGGUGCAGCAUUCCACAGCAAGAAAAUGUGGCAUAAAAUGCAAUACCUA ..(((((((((..((((((---(.((((.((.....)).)))).)))))))..)))))))...........((((...))))....)).. ( -34.20) >consensus UUGGGCUGUGGGUUGUUGUU__GGCUGCCGUUGCAAGUUGCAGGUGCAGCAUUCCACAGAAAGAAAAUAUGUCAGAAAAUUCAAUAUCUA .....((((((..((((((....((.((........)).))....))))))..))))))............................... (-21.52 = -21.04 + -0.48)



| Location | 24,171,805 – 24,171,895 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 90 |
| Reading direction | reverse |
| Mean pairwise identity | 86.93 |
| Mean single sequence MFE | -19.73 |
| Consensus MFE | -14.72 |
| Energy contribution | -15.12 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.779854 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24171805 90 - 27905053 UAGAUAUAGAUAUUUCUUACAAAAUUUCUUUCUGUGGAAUGCUGCACCUGCCGCUUGCAACGGCAGCCACAACAACAACCCACAGCCUAA (((....(((.((((......)))).)))..((((((..((.((...((((((.......))))))......)).))..)))))).))). ( -19.20) >DroSec_CAF1 12417 89 - 1 AAGAUUUGGAAUUUUCUGACAUAUUUUCUUUCUGUGGAAUGCUGCACCUGCAACUUGCAACGGCAGCC-UAACAACAACCCACAGCCCAA .....(..((....))..)............((((((...(((((...(((.....)))...))))).-..........))))))..... ( -18.34) >DroSim_CAF1 12357 90 - 1 UAGAUUUAGAAUUUUCUGACAUAUUUUCUUUCUGUGGAAUGCUGCACCUGCAACUUGCAACGGCAGCCGCAACAACAACCCACAGCCCAA .....(((((....)))))............((((((...(((((...(((.....)))...))))).(......)...))))))..... ( -20.40) >DroEre_CAF1 5686 87 - 1 UAGAUAUUGCAUUUUAUGCCACAUUUGUUUUCUGUGGAAUGCUGCACCUGCAACUUGCAACGACAGCC---ACAACAACCCACAGCCCAA (((((...((((...))))...)))))....((((((...((((....(((.....)))....)))).---........))))))..... ( -14.70) >DroYak_CAF1 5942 87 - 1 UAGGUAUUGCAUUUUAUGCCACAUUUUCUUGCUGUGGAAUGCUGCACCUGCAGCUUGCAACGACAGCC---ACAACAACCCACAGCCCAA ..(((((........)))))..........(((((((...(((((....)))))..((.......)).---........))))))).... ( -26.00) >consensus UAGAUAUAGAAUUUUCUGACACAUUUUCUUUCUGUGGAAUGCUGCACCUGCAACUUGCAACGGCAGCC__AACAACAACCCACAGCCCAA ...............................((((((...(((((...(((.....)))...)))))............))))))..... (-14.72 = -15.12 + 0.40)


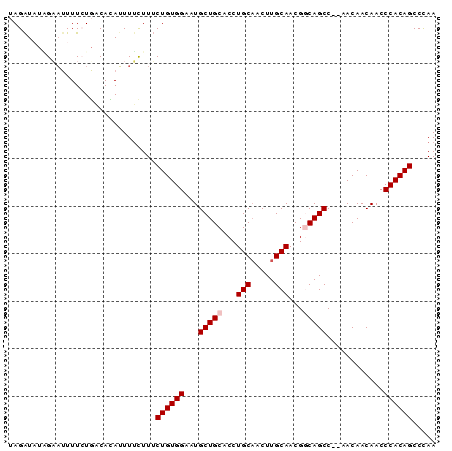
| Location | 24,171,821 – 24,171,935 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 76.54 |
| Mean single sequence MFE | -29.15 |
| Consensus MFE | -14.69 |
| Energy contribution | -13.93 |
| Covariance contribution | -0.76 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.46 |
| Structure conservation index | 0.50 |
| SVM decision value | 2.05 |
| SVM RNA-class probability | 0.986736 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24171821 114 + 27905053 UGUUGUGGCUGCCGUUGCAAGCGGCAGGUGCAGCAUUCCACAGAAAGAAAUUUUGUAAGAAAUAUCUAUAUCUAGAAGGUGAGUAUAUAAUGCAUUAUUAUGCAUGACACCAAG (((((((.(((((((.....))))))).)))))))(((.((((((.....))))))..)))...((((....)))).((((........((((((....))))))..))))... ( -34.50) >DroSec_CAF1 12433 110 + 1 UGUUA-GGCUGCCGUUGCAAGUUGCAGGUGCAGCAUUCCACAGAAAGAAAAUAUGUCAGAAAAUUCCAAAUCUUGAACGGGAGUAUAUUAUGCAUA---CGGAAUGACACAAAA (((((-.....((((((((.(((((....)))))(((((.....((((......................)))).....)))))......)))).)---)))..)))))..... ( -25.25) >DroSim_CAF1 12373 111 + 1 UGUUGCGGCUGCCGUUGCAAGUUGCAGGUGCAGCAUUCCACAGAAAGAAAAUAUGUCAGAAAAUUCUAAAUCUAGAAUGGGAGUAUAUGAUGCAUA---CGGAAUGACACAAAA ..((((((((((....)).))))))))(((...((((((................((.....((((((....))))))..))((((.......)))---))))))).))).... ( -25.90) >DroEre_CAF1 5702 90 + 1 UGU---GGCUGUCGUUGCAAGUUGCAGGUGCAGCAUUCCACAGAAAACAAAUGUGGCAUAAAAUGCAAUAUCUAGA------------------UA---UGGAUUGCUACAAAA (((---(((....((((((.(((((....)))))...(((((.........))))).......))))))(((((..------------------..---))))).))))))... ( -27.60) >DroYak_CAF1 5958 108 + 1 UGU---GGCUGUCGUUGCAAGCUGCAGGUGCAGCAUUCCACAGCAAGAAAAUGUGGCAUAAAAUGCAAUACCUAGAAAGGGAGUAUAUUAUGGACA---UGGAAUGCUACAAAA (((---(((((((((((((.(((((....)))))...(((((.........))))).......)))))).(((....)))............))))---......))))))... ( -32.50) >consensus UGUU__GGCUGCCGUUGCAAGUUGCAGGUGCAGCAUUCCACAGAAAGAAAAUAUGUCAGAAAAUUCAAUAUCUAGAA_GGGAGUAUAUUAUGCAUA___CGGAAUGACACAAAA (((....(((((..((((.....))))..))))).....)))...........(((((..............................................)))))..... (-14.69 = -13.93 + -0.76)

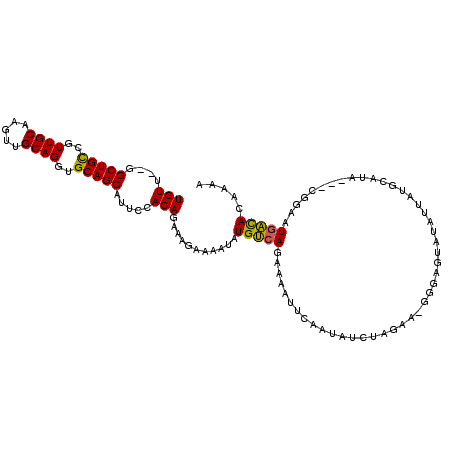
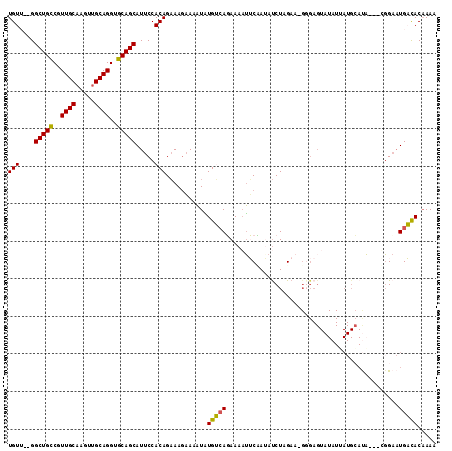
| Location | 24,171,821 – 24,171,935 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 76.54 |
| Mean single sequence MFE | -23.96 |
| Consensus MFE | -10.77 |
| Energy contribution | -11.53 |
| Covariance contribution | 0.76 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.45 |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.768978 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24171821 114 - 27905053 CUUGGUGUCAUGCAUAAUAAUGCAUUAUAUACUCACCUUCUAGAUAUAGAUAUUUCUUACAAAAUUUCUUUCUGUGGAAUGCUGCACCUGCCGCUUGCAACGGCAGCCACAACA ....((((.((((((....)))))).)))).....((...((((...(((.((((......)))).))).)))).))...(((((...(((.....)))...)))))....... ( -24.00) >DroSec_CAF1 12433 110 - 1 UUUUGUGUCAUUCCG---UAUGCAUAAUAUACUCCCGUUCAAGAUUUGGAAUUUUCUGACAUAUUUUCUUUCUGUGGAAUGCUGCACCUGCAACUUGCAACGGCAGCC-UAACA ...((((((((((((---((((.....)))))...(......)....)))).....)))))))..((((......)))).(((((...(((.....)))...))))).-..... ( -23.00) >DroSim_CAF1 12373 111 - 1 UUUUGUGUCAUUCCG---UAUGCAUCAUAUACUCCCAUUCUAGAUUUAGAAUUUUCUGACAUAUUUUCUUUCUGUGGAAUGCUGCACCUGCAACUUGCAACGGCAGCCGCAACA ..(((((..((((((---((((.....)))))........((((...((((..(........)..)))).)))).)))))(((((...(((.....)))...)))))))))).. ( -25.20) >DroEre_CAF1 5702 90 - 1 UUUUGUAGCAAUCCA---UA------------------UCUAGAUAUUGCAUUUUAUGCCACAUUUGUUUUCUGUGGAAUGCUGCACCUGCAACUUGCAACGACAGCC---ACA ...(((((((.((((---((------------------...(((((..((((...))))......)))))..)))))).)))))))..(((.....))).........---... ( -21.40) >DroYak_CAF1 5958 108 - 1 UUUUGUAGCAUUCCA---UGUCCAUAAUAUACUCCCUUUCUAGGUAUUGCAUUUUAUGCCACAUUUUCUUGCUGUGGAAUGCUGCACCUGCAGCUUGCAACGACAGCC---ACA ...((((((((((((---(.......................(((((........))))).((......))..)))))))))))))..(((.....))).........---... ( -26.20) >consensus UUUUGUGUCAUUCCA___UAUGCAUAAUAUACUCCC_UUCUAGAUAUAGAAUUUUCUGACACAUUUUCUUUCUGUGGAAUGCUGCACCUGCAACUUGCAACGGCAGCC__AACA ...(((((((..............................................)))))))..........((.....(((((...(((.....)))...)))))....)). (-10.77 = -11.53 + 0.76)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:24:58 2006