
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
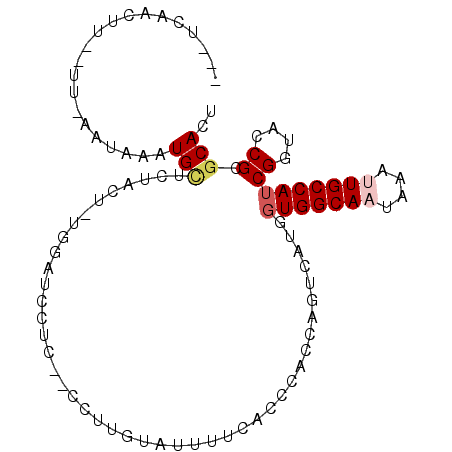
| Location | 3,042,612 – 3,042,704 |
| Length | 92 |
| Max. P | 0.830311 |

| Location | 3,042,612 – 3,042,704 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 71.46 |
| Mean single sequence MFE | -22.69 |
| Consensus MFE | -7.76 |
| Energy contribution | -8.32 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.34 |
| SVM decision value | 0.71 |
| SVM RNA-class probability | 0.830311 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3042612 92 + 27905053 ---UCAACUU--UU-AUUAAAUGCUCUACUGUGGCUCCUU--CCUUCUAUUUUCACCCACCAGUCAUGGUGGCAAUAAAUUGCCAUCGGUACCGCACACU ---.......--..-......(((..(((((((((.....--......((((....((((((....))))))....)))).)))).)))))..))).... ( -22.30) >DroPse_CAF1 30315 96 + 1 UAGUUAGUUA--UU-GAUUAGUGCGUGUUU-CAGAUGGACAACCGAAUUGAUUUACCCACCCGUCAUUGUGGCAAUAAGCUGCCAUCGAUAGCGCGCACU ..........--..-....(((((((((((-(.(((((......................)))))...((((((......)))))).)).)))))))))) ( -27.55) >DroSec_CAF1 29663 95 + 1 ---UCAACUUUUUU-AUUAAAUGCUCUACUGUGGUUCCUU-GUCUUGUAUUUUCACCCACCAGUCAUGGUGGCAAUAAAUUGCCAUCGGUACCGCGCACU ---...........-...............(((((.((.(-(.((.((..........)).)).)).((((((((....)))))))))).)))))..... ( -21.40) >DroSim_CAF1 29901 92 + 1 ---UCAACUU--UU-AUUAAAUGCUCUACUGUGGUUCCUU--CCUUGUAUUUUCACCCACCAGUCAUGGUGGCAAUAAAUUGCCAUCGGUACCGCGCACU ---.......--..-.......((..(((((((((.....--......((((....((((((....))))))....)))).)))).)))))..))..... ( -20.00) >DroAna_CAF1 32289 89 + 1 ---UAUGCUU--UUAAAUAAAUGUAUU----UAGGUCAUC--CACUGAAUCUUCACCCACCCGUCAUAGUGGCAAUAAUUUGCCAACGGUAUCGCGCACU ---..(((..--............(((----(((......--..))))))..........((((.....((((((....))))))))))......))).. ( -16.80) >DroPer_CAF1 30371 96 + 1 UAGUUAGUUA--UU-GAUUAGUGCGUGUUU-UUGAUGGACAACCGAAUAGAUUUACCCACCCGUCAUUGUGGCAAUAAGCUGCCAUCGAUAGCGCGCACU ..........--..-....((((((((((.-(((((((.((................(((........)))((.....))))))))))).)))))))))) ( -28.10) >consensus ___UCAACUU__UU_AAUAAAUGCUCUACU_UGGAUCCUC__CCUUGUAUUUUCACCCACCAGUCAUGGUGGCAAUAAAUUGCCAUCGGUACCGCGCACU .....................(((............................................(((((((....)))))))((....)).))).. ( -7.76 = -8.32 + 0.56)

| Location | 3,042,612 – 3,042,704 |
|---|---|
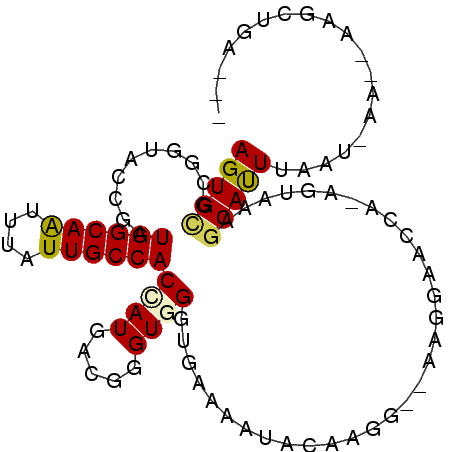
| Length | 92 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 71.46 |
| Mean single sequence MFE | -23.18 |
| Consensus MFE | -11.92 |
| Energy contribution | -11.78 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.51 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3042612 92 - 27905053 AGUGUGCGGUACCGAUGGCAAUUUAUUGCCACCAUGACUGGUGGGUGAAAAUAGAAGG--AAGGAGCCACAGUAGAGCAUUUAAU-AA--AAGUUGA--- ...((((..(((.(.((((.........((((((....))))))((....))......--.....))))).)))..)))).....-..--.......--- ( -22.50) >DroPse_CAF1 30315 96 - 1 AGUGCGCGCUAUCGAUGGCAGCUUAUUGCCACAAUGACGGGUGGGUAAAUCAAUUCGGUUGUCCAUCUG-AAACACGCACUAAUC-AA--UAACUAACUA ((((((.(.....(.((((((....))))))).....(((((((..((.((.....))))..)))))))-...).))))))....-..--.......... ( -27.00) >DroSec_CAF1 29663 95 - 1 AGUGCGCGGUACCGAUGGCAAUUUAUUGCCACCAUGACUGGUGGGUGAAAAUACAAGAC-AAGGAACCACAGUAGAGCAUUUAAU-AAAAAAGUUGA--- (((((((.((....)).))..(((((((((((((....))))))((....)).......-.........))))))))))))....-...........--- ( -19.90) >DroSim_CAF1 29901 92 - 1 AGUGCGCGGUACCGAUGGCAAUUUAUUGCCACCAUGACUGGUGGGUGAAAAUACAAGG--AAGGAACCACAGUAGAGCAUUUAAU-AA--AAGUUGA--- (((((((.((....)).))..(((((((((((((....))))))............((--......)).))))))))))))....-..--.......--- ( -21.50) >DroAna_CAF1 32289 89 - 1 AGUGCGCGAUACCGUUGGCAAAUUAUUGCCACUAUGACGGGUGGGUGAAGAUUCAGUG--GAUGACCUA----AAUACAUUUAUUUAA--AAGCAUA--- .((((......((((((((((....)))))).....)))).((((((((........(--(....))..----......)))))))).--..)))).--- ( -19.89) >DroPer_CAF1 30371 96 - 1 AGUGCGCGCUAUCGAUGGCAGCUUAUUGCCACAAUGACGGGUGGGUAAAUCUAUUCGGUUGUCCAUCAA-AAACACGCACUAAUC-AA--UAACUAACUA ((((((.(.....(((((((((((((((...))))))((((((((....)))))))))))).)))))..-...).))))))....-..--.......... ( -28.30) >consensus AGUGCGCGGUACCGAUGGCAAUUUAUUGCCACCAUGACGGGUGGGUGAAAAUACAAGG__AAGGAACCA_AGUAAAGCAUUUAAU_AA__AAGCUGA___ (((((..........((((((....))))))((((.....))))................................)))))................... (-11.92 = -11.78 + -0.14)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:55:02 2006