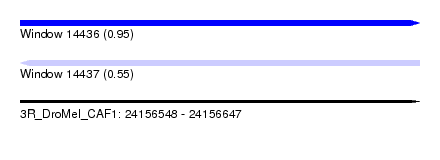
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 24,156,548 – 24,156,647 |
| Length | 99 |
| Max. P | 0.948888 |

| Location | 24,156,548 – 24,156,647 |
|---|---|
| Length | 99 |
| Sequences | 3 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 81.61 |
| Mean single sequence MFE | -35.53 |
| Consensus MFE | -25.39 |
| Energy contribution | -25.17 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.42 |
| Structure conservation index | 0.71 |
| SVM decision value | 1.39 |
| SVM RNA-class probability | 0.948888 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24156548 99 + 27905053 AAAGAGAGUGAG----GGAGAGAGGGAGCGUGCGAGAGAGUGCGGUUUCUUAUCACUCUCUGCGAAUACGUAGCCCUACGUUACGUAGCGC-CCGCUACGUUAG ..(((((((((.----.....(((..(.((..(......)..)).)..))).)))))))))......(((((....))))).(((((((..-..)))))))... ( -38.60) >DroEre_CAF1 10033 92 + 1 AAAGAGAUAGAGAG----A--------GCGCUUGAGAGAGUGCGGUUCCUUAUCACUCUCUGCGAAUACGUAGCCCUACGUUACGUAGCGCACAGCAACGUUAC ........((((((----.--------((((((....))))))(((.....))).))))))(((..((((((((.....)))))))).)))............. ( -31.70) >DroYak_CAF1 13994 103 + 1 AAAGAGAGAGAGCGCGAGAGAUAGCACGCGCGAGAGAGAGUGCGGUUUCUUAUCACUCUCUGC-AAUACGUAGCCCUACGUUACGUAGCGCACCGCAACGUUAG ..((((((.((....(((((((.((((.(.(....).).)))).))))))).)).))))))((-..((((((((.....))))))))..))............. ( -36.30) >consensus AAAGAGAGAGAG_G___GAGA_AG___GCGCGAGAGAGAGUGCGGUUUCUUAUCACUCUCUGCGAAUACGUAGCCCUACGUUACGUAGCGCACCGCAACGUUAG ...........................((((...((((((((...........)))))))).....((((((((.....))))))))))))............. (-25.39 = -25.17 + -0.22)

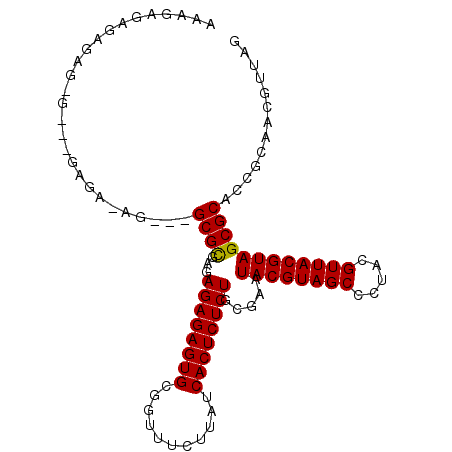
| Location | 24,156,548 – 24,156,647 |
|---|---|
| Length | 99 |
| Sequences | 3 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 81.61 |
| Mean single sequence MFE | -29.77 |
| Consensus MFE | -21.78 |
| Energy contribution | -21.67 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.554859 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24156548 99 - 27905053 CUAACGUAGCGG-GCGCUACGUAACGUAGGGCUACGUAUUCGCAGAGAGUGAUAAGAAACCGCACUCUCUCGCACGCUCCCUCUCUCC----CUCACUCUCUUU ...(((((((..-..)))))))...(.((((...(((...((.((((((((....(....).)))))))))).)))..))))).....----............ ( -29.40) >DroEre_CAF1 10033 92 - 1 GUAACGUUGCUGUGCGCUACGUAACGUAGGGCUACGUAUUCGCAGAGAGUGAUAAGGAACCGCACUCUCUCAAGCGC--------U----CUCUCUAUCUCUUU ...((((.(((.((((........)))).))).))))...(((((((((((....(....).))))))))...))).--------.----.............. ( -27.70) >DroYak_CAF1 13994 103 - 1 CUAACGUUGCGGUGCGCUACGUAACGUAGGGCUACGUAUU-GCAGAGAGUGAUAAGAAACCGCACUCUCUCUCGCGCGUGCUAUCUCUCGCGCUCUCUCUCUUU ((.((((((((........)))))))).))((........-))((((((((....(....).))))))))...(((((.(......).)))))........... ( -32.20) >consensus CUAACGUUGCGGUGCGCUACGUAACGUAGGGCUACGUAUUCGCAGAGAGUGAUAAGAAACCGCACUCUCUCACGCGC___CU_UCUC___C_CUCUCUCUCUUU ........(((..((.(((((...))))).)).........(.((((((((....(....).))))))))).)))............................. (-21.78 = -21.67 + -0.11)

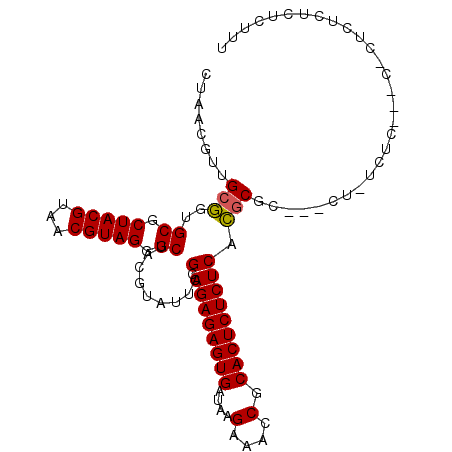
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:24:43 2006