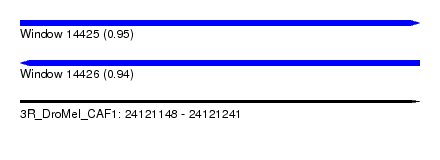
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 24,121,148 – 24,121,241 |
| Length | 93 |
| Max. P | 0.954671 |

| Location | 24,121,148 – 24,121,241 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 80.07 |
| Mean single sequence MFE | -28.57 |
| Consensus MFE | -16.78 |
| Energy contribution | -18.48 |
| Covariance contribution | 1.70 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.93 |
| Structure conservation index | 0.59 |
| SVM decision value | 1.45 |
| SVM RNA-class probability | 0.954671 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
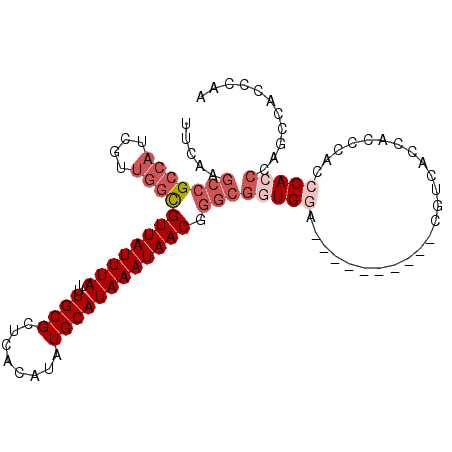
>3R_DroMel_CAF1 24121148 93 + 27905053 UUCAAGCCGCCAUCGUUGCCGUUAUUUAUUUGCGCUCACAUAUGCAUAAAUAACGGGCGGUGGA----------CGUCACCACCCACCCACCCAGCCACCCAA .....(.(((((((((..((((((((((..((((........))))))))))))))))))))).----------)).)......................... ( -25.30) >DroPse_CAF1 328130 88 + 1 -------CGCCACAGUCUGUGUUAUUUAUUUGCGCUCACAUAUGCAUAAAUAACGUGGACUGAGAGGGGGAGGGCGAGGCGCCCGAUCCACCCC-----C--- -------.....(((((..(((((((((..((((........)))))))))))))..)))))...(((((.(((((...)))))......))))-----)--- ( -37.60) >DroSec_CAF1 308151 89 + 1 UUCAAGCCGCCAUCGUUGGCGUUAUUUAUUUGCGCUCACAUAUGCAUAAAUAACGGGCGGUGGA----------CGUCACCACC----CACCAAGCCACCCAA .....(((((((....))))((((((((..((((........)))))))))))).)))(((((.----------.........)----))))........... ( -28.10) >DroSim_CAF1 308300 93 + 1 UUCAAGCCGCCAUCGUUGGCGUUAUUUAUUUGCGCUCACAUAUGCAUAAAUAACGGGCGGUGGA----------CGUCACCACCCACCCACCAAGUCACCCAA .....(((((((....))))((((((((..((((........)))))))))))).)))(((((.----------.....)))))................... ( -27.90) >DroEre_CAF1 323644 93 + 1 UUCCAGCCGCCAUCGUUGGCGUUAUUUAUUUGCGCUCACAUAUGCAUAAAUAACGGGCGCUGGA----------CGUCACCACCCACCCAUCGAACCACCCAA .(((((((((((....))))((((((((..((((........))))))))))))..).))))))----------............................. ( -24.60) >DroYak_CAF1 322300 93 + 1 UUUAAGCCGCCAUCGUUGGCGUUAUUUAUUUGCGCUCACAUAUGCAUAAAUAACGGGCGGUGGA----------CGUCACCACCCACCCACCCGACCACCCAG .....(((((((....))))((((((((..((((........)))))))))))).)))(((((.----------.....)))))................... ( -27.90) >consensus UUCAAGCCGCCAUCGUUGGCGUUAUUUAUUUGCGCUCACAUAUGCAUAAAUAACGGGCGGUGGA__________CGUCACCACCCACCCACCCAGCCACCCAA .....(((((((....))))((((((((..((((........)))))))))))).)))(((((........................)))))........... (-16.78 = -18.48 + 1.70)


| Location | 24,121,148 – 24,121,241 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 80.07 |
| Mean single sequence MFE | -35.50 |
| Consensus MFE | -19.35 |
| Energy contribution | -21.76 |
| Covariance contribution | 2.41 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.85 |
| Structure conservation index | 0.55 |
| SVM decision value | 1.25 |
| SVM RNA-class probability | 0.936449 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
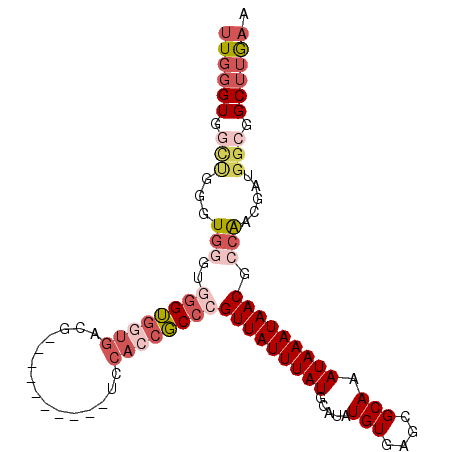
>3R_DroMel_CAF1 24121148 93 - 27905053 UUGGGUGGCUGGGUGGGUGGGUGGUGACG----------UCCACCGCCCGUUAUUUAUGCAUAUGUGAGCGCAAAUAAAUAACGGCAACGAUGGCGGCUUGAA (..(((.(((((((((.(((..(....).----------.)))))))))(((((((((.....(((....))).)))))))))(....)...))).)))..). ( -40.10) >DroPse_CAF1 328130 88 - 1 ---G-----GGGGUGGAUCGGGCGCCUCGCCCUCCCCCUCUCAGUCCACGUUAUUUAUGCAUAUGUGAGCGCAAAUAAAUAACACAGACUGUGGCG------- ---(-----((((.(((..(((((...))))))))))))).(((((...(((((((((.....(((....))).)))))))))...))))).....------- ( -33.10) >DroSec_CAF1 308151 89 - 1 UUGGGUGGCUUGGUG----GGUGGUGACG----------UCCACCGCCCGUUAUUUAUGCAUAUGUGAGCGCAAAUAAAUAACGCCAACGAUGGCGGCUUGAA (..(((.((((((((----(((((((...----------..))))))))(((((((((.....(((....))).))))))))))))).....))).)))..). ( -36.40) >DroSim_CAF1 308300 93 - 1 UUGGGUGACUUGGUGGGUGGGUGGUGACG----------UCCACCGCCCGUUAUUUAUGCAUAUGUGAGCGCAAAUAAAUAACGCCAACGAUGGCGGCUUGAA ..((....)).....(((((..(....).----------.)))))(((.(((((((((.....(((....))).)))))))))((((....)))))))..... ( -34.30) >DroEre_CAF1 323644 93 - 1 UUGGGUGGUUCGAUGGGUGGGUGGUGACG----------UCCAGCGCCCGUUAUUUAUGCAUAUGUGAGCGCAAAUAAAUAACGCCAACGAUGGCGGCUGGAA ((.(((.(((((.(((..(((((.((...----------..)).)))))(((((((((.....(((....))).))))))))).))).)))..)).))).)). ( -31.10) >DroYak_CAF1 322300 93 - 1 CUGGGUGGUCGGGUGGGUGGGUGGUGACG----------UCCACCGCCCGUUAUUUAUGCAUAUGUGAGCGCAAAUAAAUAACGCCAACGAUGGCGGCUUAAA .(((((.(((((((((.(((..(....).----------.)))))))))(((((((((.....(((....))).))))))))).........))).))))).. ( -38.00) >consensus UUGGGUGGCUGGGUGGGUGGGUGGUGACG__________UCCACCGCCCGUUAUUUAUGCAUAUGUGAGCGCAAAUAAAUAACGCCAACGAUGGCGGCUUGAA ((((((.(((...(((..((((((((...............))))))))(((((((((.....(((....))).))))))))).))).....))).)))))). (-19.35 = -21.76 + 2.41)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:24:33 2006