| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 561,330 – 561,443 |
| Length | 113 |
| Max. P | 0.606274 |

| Location | 561,330 – 561,443 |
|---|---|
| Length | 113 |
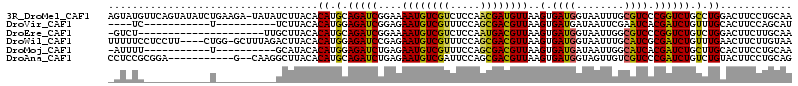
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 71.96 |
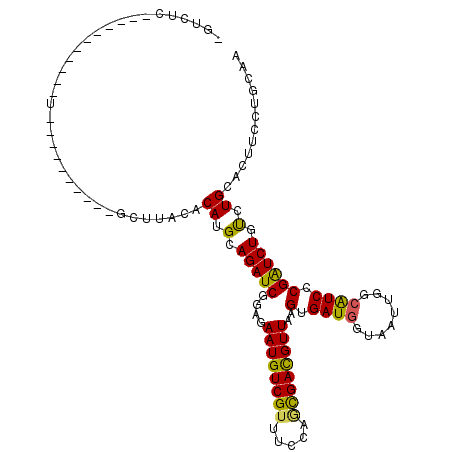
| Mean single sequence MFE | -27.27 |
| Consensus MFE | -16.50 |
| Energy contribution | -15.70 |
| Covariance contribution | -0.80 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.26 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.606274 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 561330 113 + 27905053 AGUAUGUUCAGUAUAUCUGAAGA-UAUAUCUUACACAUGCAGAUCGGAAAAUGUCGUCUCCAACGAUGUUAAGUGAUGGUAAUUUGCGUCCCGGUCUGCCUGGACUUCCUGCAA .(((.((((((((((((....))-))))..........(((((((((..((((((((.....))))))))....((((........)))))))))))))))))))....))).. ( -34.70) >DroVir_CAF1 7246 89 + 1 ----UC-----------U----------UCUUACACAUGGAGAUCGGAGAAUGUCGUUUCCAGCGACGUUAAGUGAUGAUAAUUCGAAUCACGAUCUGUUUGCACUUCCAGCAU ----((-----------(----------((........)))))..(((.((((((((.....)))))))).((((..((((..(((.....)))..))))..)))))))..... ( -23.30) >DroEre_CAF1 447 92 + 1 -GUCU---------------------UUGCUUACACAUGCAGAUCGGAAAAUGUCGUCUCCAAUGACGUUAAGUGAUGGUAAUUGGCGUCCCGGUCUGUCUGGACUUCUUGCAA -....---------------------((((.....((.(((((((((..((((((((.....))))))))....((((.(....).))))))))))))).))........)))) ( -26.62) >DroWil_CAF1 458 109 + 1 UUUUUCCUCCUU----CUGG-GCUUUAGACUUACACAUGGAGAUCCGAGAAUGUCGUUUCCAGCGACGUUAAGUGAUGGUAAUUUGCAUCGCGAUCUGUUUGAACUUCUUGUAA ............----..((-(.(((((((...(....).(((((....((((((((.....))))))))..((((((........)))))))))))))))))).)))...... ( -24.50) >DroMoj_CAF1 482 92 + 1 -AUUUU-----------U----------GCAUACACAUGGAGAUCUGAGAAUGUCGUUUCCAGCGACGUUAAGUGAUGAUAAUUGGCAUCACGAUCUGCUUGCACUUCCUGCAA -....(-----------(----------(((....((.(.(((((....((((((((.....))))))))..((((((........))))))))))).).)).......))))) ( -26.60) >DroAna_CAF1 470 101 + 1 CCUCCGCGGA-----------G--CAAGGCUUACACAUGCAGAUCUGAGAAUGUCGAUUCCAGCGACGUUAAGUGAUGGUAGUUGUCGUCCCGAUCUGUCUGUACUUCCUGCAG .....(((((-----------(--(...)))...(((.(((((((....(((((((.......)))))))..(.(((((......))))).)))))))).))).....)))).. ( -27.90) >consensus _GUCUC___________U__________GCUUACACAUGCAGAUCGGAGAAUGUCGUUUCCAGCGACGUUAAGUGAUGGUAAUUGGCAUCCCGAUCUGUCUGCACUUCCUGCAA ...................................((.(.(((((....((((((((.....))))))))..(.((((........)))).)))))).).))............ (-16.50 = -15.70 + -0.80)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:32:47 2006