
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 24,097,233 – 24,097,326 |
| Length | 93 |
| Max. P | 0.753444 |

| Location | 24,097,233 – 24,097,326 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 93 |
| Reading direction | forward |
| Mean pairwise identity | 81.62 |
| Mean single sequence MFE | -27.92 |
| Consensus MFE | -23.13 |
| Energy contribution | -23.27 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.753444 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
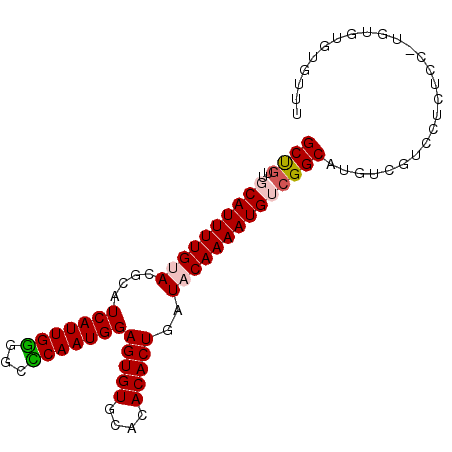
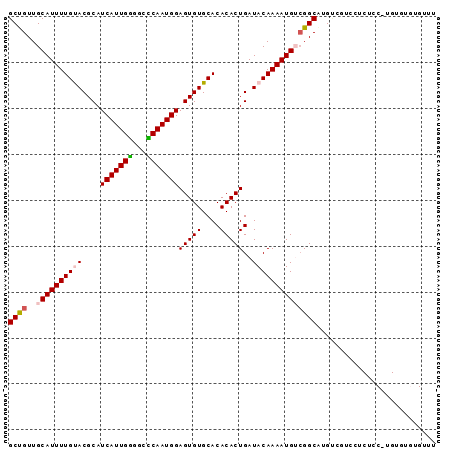
>3R_DroMel_CAF1 24097233 93 + 27905053 GCUGUUGCAUUUUGUACGCAUCAUUGGGGCCCAAUGGAGUGUGCACACACUGAUACAAAAUGUCGGCGUGUCGUCCUCUCCUUGUGUGUGUUU ((....))......(((((((.(((((...)))))((((.(..(((((.((((((.....)))))).)))).)..).))))..)))))))... ( -29.00) >DroVir_CAF1 305649 80 + 1 GCCGUUGCAUUUUGCACGCAUCAUUGAUGCUCAAUGGAGUGUGCACACACUGAUACAAAAUGGCGGCAGGACAUG---------CGAGU---- (((((..((((((((((((..(((((.....)))))..)))))).(.....)....)))))))))))........---------.....---- ( -26.10) >DroGri_CAF1 263402 80 + 1 GCCGUUGCAUUUUGCACGCAUCAUUGAUGCUCAAUGGAGUGUGCACACACUGAUACAAAAUGGCGGCAGGCCAUA---------UGAGA---- (((((((((((((((((((..(((((.....)))))..)))))).(.....)....))))).))))).)))....---------.....---- ( -26.20) >DroSec_CAF1 284495 93 + 1 GCUGUUGCAUUUUGUACGCAUCAUUGGGGCCCAAUGGAGUGUGCACACACUGAUACAAAAUGUAGGCGUGUCGUCCUCUCCAUGUGUGUGUUU ((....)).....((((((..((((((...))))))..))))))((((((...(((.....))).(((((..(....)..))))))))))).. ( -28.40) >DroSim_CAF1 284406 93 + 1 GCUGUUGCAUUUUGUACGCAUCAUUGGGGCCCAAUGGAGUGUGCACACACUGAUACAAAAUGUCGGCGUGUCGUCCUCUCCAUGUGUGUGUUU ((....))......(((((((...(((...)))((((((.(..(((((.((((((.....)))))).)))).)..).)))))))))))))... ( -29.50) >DroAna_CAF1 293533 93 + 1 GCUGUUGCAUUUUGUACGCAUCAUUGGGGCUCAAUGGAGUGUGCACACACUGAUACAAAAUGUCGGCAUGUCUUUCGCUUCGUGUAUGCGACG ...(((((((...((((((..(((((.....)))))..))))))((((.((((((.....))))))...((.....))...))))))))))). ( -28.30) >consensus GCUGUUGCAUUUUGUACGCAUCAUUGGGGCCCAAUGGAGUGUGCACACACUGAUACAAAAUGUCGGCAUGUCGUCCUCUCC_UGUGUGUGUUU ((((..((((((((((....(((((((...)))))))(((((....)))))..)))))))))))))).......................... (-23.13 = -23.27 + 0.14)

| Location | 24,097,233 – 24,097,326 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 81.62 |
| Mean single sequence MFE | -22.97 |
| Consensus MFE | -17.75 |
| Energy contribution | -17.53 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.32 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.595186 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
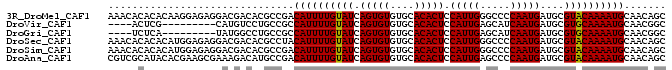
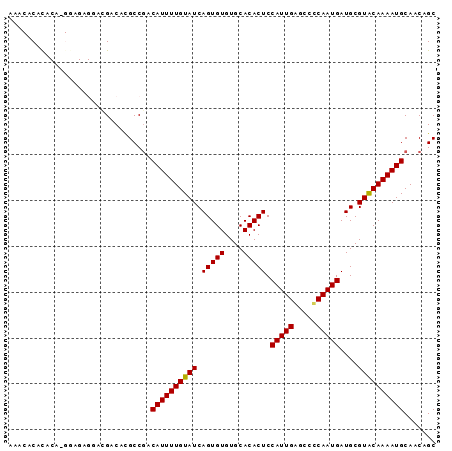
>3R_DroMel_CAF1 24097233 93 - 27905053 AAACACACACAAGGAGAGGACGACACGCCGACAUUUUGUAUCAGUGUGUGCACACUCCAUUGGGCCCCAAUGAUGCGUACAAAAUGCAACAGC ............((.(.(.....).).))(.((((((((((.(((((....))))).((((((...))))))....)))))))))))...... ( -20.80) >DroVir_CAF1 305649 80 - 1 ----ACUCG---------CAUGUCCUGCCGCCAUUUUGUAUCAGUGUGUGCACACUCCAUUGAGCAUCAAUGAUGCGUGCAAAAUGCAACGGC ----.....---------........((((.(((((((((((((((.((....))..))))))(((((...))))).)))))))))...)))) ( -24.10) >DroGri_CAF1 263402 80 - 1 ----UCUCA---------UAUGGCCUGCCGCCAUUUUGUAUCAGUGUGUGCACACUCCAUUGAGCAUCAAUGAUGCGUGCAAAAUGCAACGGC ----.....---------........((((.(((((((((((((((.((....))..))))))(((((...))))).)))))))))...)))) ( -24.10) >DroSec_CAF1 284495 93 - 1 AAACACACACAUGGAGAGGACGACACGCCUACAUUUUGUAUCAGUGUGUGCACACUCCAUUGGGCCCCAAUGAUGCGUACAAAAUGCAACAGC .......((.((((((.(..(.((((((.(((.....)))...)))))))..).))))))))........((.(((((.....))))).)).. ( -22.20) >DroSim_CAF1 284406 93 - 1 AAACACACACAUGGAGAGGACGACACGCCGACAUUUUGUAUCAGUGUGUGCACACUCCAUUGGGCCCCAAUGAUGCGUACAAAAUGCAACAGC .......((.((((((.(..(.((((((..((.....))....)))))))..).))))))))........((.(((((.....))))).)).. ( -21.70) >DroAna_CAF1 293533 93 - 1 CGUCGCAUACACGAAGCGAAAGACAUGCCGACAUUUUGUAUCAGUGUGUGCACACUCCAUUGAGCCCCAAUGAUGCGUACAAAAUGCAACAGC .((.((((((((((.((((((............)))))).)).))))))))))....(((((.....))))).(((((.....)))))..... ( -24.90) >consensus AAACACACACA_GGAGAGGACGACACGCCGACAUUUUGUAUCAGUGUGUGCACACUCCAUUGAGCCCCAAUGAUGCGUACAAAAUGCAACAGC ...............................((((((((((.(((((....))))).(((((.....)))))....))))))))))....... (-17.75 = -17.53 + -0.22)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:24:15 2006