
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 24,093,295 – 24,093,431 |
| Length | 136 |
| Max. P | 0.848488 |

| Location | 24,093,295 – 24,093,397 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 81.90 |
| Mean single sequence MFE | -40.84 |
| Consensus MFE | -31.92 |
| Energy contribution | -33.12 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.78 |
| SVM RNA-class probability | 0.848488 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24093295 102 + 27905053 CGGCUCCACAUGUAGUACAUCCUGCAUGAGCCAAAUGCGGCUGGCUCCUGCCAAGGAUA-------AGCUCGAGCUCAGCCUCUUGGCGCUGCAGCCGGUUGGCCUACA (((((.(((((((((......))))))).(((((..(.(((((((((..((........-------.))..)))).))))).))))))..)).)))))........... ( -41.10) >DroSec_CAF1 280624 109 + 1 CGGCUCCACAUGUAGUACAUUCUGCAUGAGUCUAAUGCGGCUGGCUCCUGCCAAGGAUGAGCAAGGAGCUCGAGCUCAGCCUCUUGGCGCUGCAGUCGGUUGGCCUGUA .((((((.(((((((......))))))).......((((((.(((((((((((....)).)).)))))))........(((....)))))))))...))..)))).... ( -44.40) >DroSim_CAF1 280337 109 + 1 GGGCUCCACAUGUAGUACAUUCUGCAUGAGUCUAAUGCGGCUGGCUCCUGCCAAGGAUGAACAGGGAGCUCGAGCUCAGCCUCUUGGCGCUGCAGUCGGUUGGCCUGUA (((((((.(((((((......))))))).......((((((((((....))))............((((....)))).(((....)))))))))...))..)))))... ( -42.90) >DroEre_CAF1 294032 96 + 1 UGGCUCCACAUA-----CAUUCUGUAUGAACCUAAUGCGGUUGGCCCAUGCCAAGGAUGAGCAGGG------AGCUCAGCCUCUUGGCGCUGCAGCCGGUUGGCCUG-- .((((((.((((-----(.....)))))((((......))))(((.((((((((((.(((((....------.)))))...)))))))).))..)))))..))))..-- ( -38.00) >DroYak_CAF1 292904 102 + 1 UGGCUCCACAUACA-UACAUGCUGCAAGAACCUAAUGCGGUUGCCUCAUGCCAAGGAUGAGCAGGGA----GAGCUCAGCCUCUUGGCGAUGCAACCGCUUGGCCUG-- .((((.(((((...-...))).))............((((((((.((..(((((((.(((((.....----..)))))...))))))))).))))))))..))))..-- ( -37.80) >consensus CGGCUCCACAUGUAGUACAUUCUGCAUGAGCCUAAUGCGGCUGGCUCCUGCCAAGGAUGAGCAGGGAGCUCGAGCUCAGCCUCUUGGCGCUGCAGCCGGUUGGCCUG_A .((((...(((((((......))))))).........((((((.(...((((((((.(((((...........)))))...))))))))..)))))))...)))).... (-31.92 = -33.12 + 1.20)

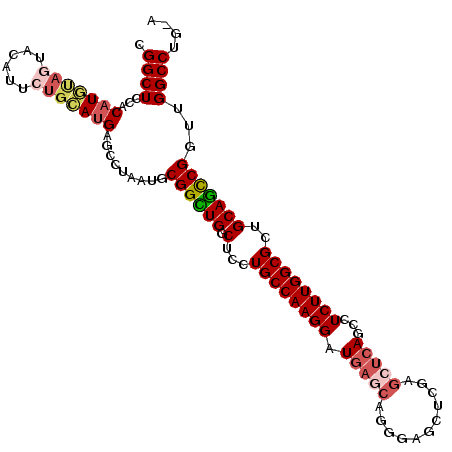
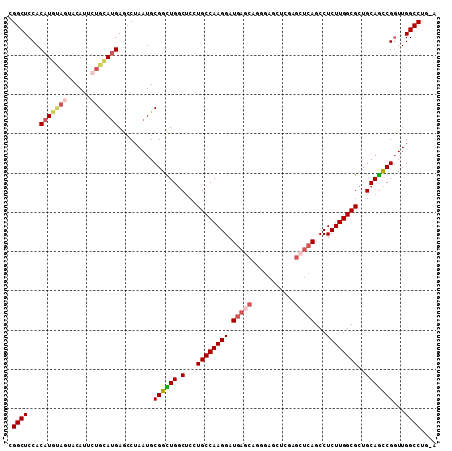
| Location | 24,093,295 – 24,093,397 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 81.90 |
| Mean single sequence MFE | -39.32 |
| Consensus MFE | -26.68 |
| Energy contribution | -27.20 |
| Covariance contribution | 0.52 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.702472 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24093295 102 - 27905053 UGUAGGCCAACCGGCUGCAGCGCCAAGAGGCUGAGCUCGAGCU-------UAUCCUUGGCAGGAGCCAGCCGCAUUUGGCUCAUGCAGGAUGUACUACAUGUGGAGCCG ....(((...(((.((((((.((((((.(((((.((((..(((-------.......)))..)))))))))...)))))).).))))).(((.....))).))).))). ( -41.80) >DroSec_CAF1 280624 109 - 1 UACAGGCCAACCGACUGCAGCGCCAAGAGGCUGAGCUCGAGCUCCUUGCUCAUCCUUGGCAGGAGCCAGCCGCAUUAGACUCAUGCAGAAUGUACUACAUGUGGAGCCG ....(((....(((((.((((........)))))).))).((((((.((.((....))))))))))...(((((((((...(((.....)))..))).)))))).))). ( -36.20) >DroSim_CAF1 280337 109 - 1 UACAGGCCAACCGACUGCAGCGCCAAGAGGCUGAGCUCGAGCUCCCUGUUCAUCCUUGGCAGGAGCCAGCCGCAUUAGACUCAUGCAGAAUGUACUACAUGUGGAGCCC .....(((((......((((.(((....))).((((....)))).))))......))))).((.((...(((((((((...(((.....)))..))).)))))).)))) ( -32.80) >DroEre_CAF1 294032 96 - 1 --CAGGCCAACCGGCUGCAGCGCCAAGAGGCUGAGCU------CCCUGCUCAUCCUUGGCAUGGGCCAACCGCAUUAGGUUCAUACAGAAUG-----UAUGUGGAGCCA --..(((.....((((.((..((((((.((.(((((.------....))))))))))))).))))))..((((((.(.((((.....)))).-----))))))).))). ( -41.50) >DroYak_CAF1 292904 102 - 1 --CAGGCCAAGCGGUUGCAUCGCCAAGAGGCUGAGCUC----UCCCUGCUCAUCCUUGGCAUGAGGCAACCGCAUUAGGUUCUUGCAGCAUGUA-UGUAUGUGGAGCCA --........((((((((.((((((((.((.(((((..----.....)))))))))))))..)).))))))))....((((((....(((((..-..))))))))))). ( -44.30) >consensus U_CAGGCCAACCGGCUGCAGCGCCAAGAGGCUGAGCUCGAGCUCCCUGCUCAUCCUUGGCAGGAGCCAGCCGCAUUAGGCUCAUGCAGAAUGUACUACAUGUGGAGCCA ....(((..............((((((.((.(((((...........))))))))))))).........((((((......(((.....)))......)))))).))). (-26.68 = -27.20 + 0.52)



| Location | 24,093,324 – 24,093,431 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 74.42 |
| Mean single sequence MFE | -40.22 |
| Consensus MFE | -19.51 |
| Energy contribution | -22.54 |
| Covariance contribution | 3.03 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.49 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.580272 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24093324 107 - 27905053 AAUGCGGCUCAUUAGGCCCGGUCAGAGUUCAGAGUGUAGGCCAACCGGCUGCAGCGCCAAGAGGCUGAGCUCGAGCU-------UAUCCUUGGCAGGAGCCAGCCGCAUUUGGC (((((((((.....(((((.((((((((((.((((...((((....(((......)))....))))..)))))))))-------).....)))).)).)))))))))))).... ( -46.20) >DroSec_CAF1 280653 114 - 1 AAUGCUUCUCAUUAGGCCCGGUCUGAGUUCAGAGUACAGGCCAACCGACUGCAGCGCCAAGAGGCUGAGCUCGAGCUCCUUGCUCAUCCUUGGCAGGAGCCAGCCGCAUUAGAC (((((.........((((.(.((((....))))...).))))....................(((((.((((((((.....))))...((....)))))))))))))))).... ( -38.70) >DroSim_CAF1 280366 114 - 1 AAUGCGGCUCAUUAGGCCCGGUCAGAGUUCAGAGUACAGGCCAACCGACUGCAGCGCCAAGAGGCUGAGCUCGAGCUCCCUGUUCAUCCUUGGCAGGAGCCAGCCGCAUUAGAC (((((((((.....(((((.(((((((((((.(((...((....)).))).....(((....))))))))))((((.....)))).....)))).)).)))))))))))).... ( -45.80) >DroEre_CAF1 294056 103 - 1 AAUGCGCCUCAUUAGGUCCGGUCAGAGUUCA-----CAGGCCAACCGGCUGCAGCGCCAAGAGGCUGAGCU------CCCUGCUCAUCCUUGGCAUGGGCCAACCGCAUUAGGU ((((((........(((..((((........-----..)))).)))((((.((..((((((.((.(((((.------....))))))))))))).))))))...)))))).... ( -39.40) >DroYak_CAF1 292932 105 - 1 AAUGCGCCUCAUUAGGCCCGGUCAGAGUUCA-----CAGGCCAAGCGGUUGCAUCGCCAAGAGGCUGAGCUC----UCCCUGCUCAUCCUUGGCAUGAGGCAACCGCAUUAGGU .....((((.....((((.(...........-----).))))..((((((((.((((((((.((.(((((..----.....)))))))))))))..)).))))))))...)))) ( -44.80) >DroAna_CAF1 289885 89 - 1 AGUGCGGCUCAUUAGGCCAGGCCGGAGACCU-----CAG-----AAGGCUGCAACGCCAAAAGACAGAGGCCGAG--------------UAGGCAUU-UCCAGCAAUAUUGCAA (((((((((.....)))).(((((....)..-----...-----..(((......)))..........))))...--------------...)))))-....(((....))).. ( -26.40) >consensus AAUGCGGCUCAUUAGGCCCGGUCAGAGUUCA_____CAGGCCAACCGGCUGCAGCGCCAAGAGGCUGAGCUCGAG_UCCCUGCUCAUCCUUGGCAGGAGCCAGCCGCAUUAGAC ((((((((((....((((.((((...............))))....)))).....((((((.((.(((((...........)))))))))))))..)))))....))))).... (-19.51 = -22.54 + 3.03)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:24:06 2006