
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 24,047,620 – 24,047,782 |
| Length | 162 |
| Max. P | 0.965958 |

| Location | 24,047,620 – 24,047,711 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.41 |
| Mean single sequence MFE | -20.35 |
| Consensus MFE | -17.63 |
| Energy contribution | -18.13 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.571899 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24047620 91 + 27905053 ----------------------------GUCGGGUCCCAUGUUGAUAAUUCGAAAUUUGUAUUCCAAUCUUCAAAUUGUCU-CCAAAAUGGAAUUGAUUUUAUGCAGAUGAGCAUGAAAU ----------------------------.((((.(((..((..(((((((.(((..(((.....)))..))).))))))).-.))....))).))))((((((((......)))))))). ( -20.90) >DroVir_CAF1 265365 89 + 1 -----------------------------U-GUGUCCCACGUUGAUAAUUCGAAAUUUGUAUUCCAAUCUUCAAAUUGUCU-CCAAAAUGGAAUUGAUUUUAUGCAGAUGAGCAUGAAAU -----------------------------.-(((...)))...(((((((.(((..(((.....)))..))).)))))))(-((.....)))....(((((((((......))))))))) ( -18.40) >DroPse_CAF1 261204 91 + 1 -----------------------------GGGGGUCCCAUGUUGAUAAUUCGAAAUUUGUAUUCCAAUCUUCAAAUUGUCUACCAAAAUGGAAUUGAUUUUAUGCAGAUGAGCAUGAAAU -----------------------------(((...))).....(((((((.(((..(((.....)))..))).)))))))..((.....)).....(((((((((......))))))))) ( -19.50) >DroWil_CAF1 294303 119 + 1 UAUACUUCUUCUGGUUGUCCUCUUUUUUUUCUGGUCCCAGGUUGAUAAUUCGAAAUUUGUAUUCCAAUCUUCAAAUUGUCU-CCAAAAUGGAAUUGAUUUUAUGCAGAUGAGCAUGAAAU .........(((((..(.((............)).))))))..(((((((.(((..(((.....)))..))).)))))))(-((.....)))....(((((((((......))))))))) ( -23.90) >DroAna_CAF1 252211 89 + 1 ------------------------------UGGGUCCCCUGUUGAUAAUUCGAAAUUUGUAUUCCAAUCUUCAAAUUGUCU-CCAAAAUGGAAUUGAUUUUAUGCAGAUGAGCAUGAAAU ------------------------------.((....))....(((((((.(((..(((.....)))..))).)))))))(-((.....)))....(((((((((......))))))))) ( -19.90) >DroPer_CAF1 262608 91 + 1 -----------------------------GGGGGUCCCAUGUUGAUAAUUCGAAAUUUGUAUUCCAAUCUUCAAAUUGUCUACCAAAAUGGAAUUGAUUUUAUGCAGAUGAGCAUGAAAU -----------------------------(((...))).....(((((((.(((..(((.....)))..))).)))))))..((.....)).....(((((((((......))))))))) ( -19.50) >consensus _____________________________UCGGGUCCCAUGUUGAUAAUUCGAAAUUUGUAUUCCAAUCUUCAAAUUGUCU_CCAAAAUGGAAUUGAUUUUAUGCAGAUGAGCAUGAAAU ....................................((((...(((((((.(((..(((.....)))..))).))))))).......)))).....(((((((((......))))))))) (-17.63 = -18.13 + 0.50)



| Location | 24,047,620 – 24,047,711 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.41 |
| Mean single sequence MFE | -20.97 |
| Consensus MFE | -18.18 |
| Energy contribution | -18.40 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.63 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.59 |
| SVM RNA-class probability | 0.965958 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24047620 91 - 27905053 AUUUCAUGCUCAUCUGCAUAAAAUCAAUUCCAUUUUGG-AGACAAUUUGAAGAUUGGAAUACAAAUUUCGAAUUAUCAACAUGGGACCCGAC---------------------------- ((((.((((......)))).))))...((((((.((((-....(((((((((.(((.....))).))))))))).)))).))))))......---------------------------- ( -19.30) >DroVir_CAF1 265365 89 - 1 AUUUCAUGCUCAUCUGCAUAAAAUCAAUUCCAUUUUGG-AGACAAUUUGAAGAUUGGAAUACAAAUUUCGAAUUAUCAACGUGGGACAC-A----------------------------- ((((.((((......)))).))))...((((((.((((-....(((((((((.(((.....))).))))))))).)))).))))))...-.----------------------------- ( -19.60) >DroPse_CAF1 261204 91 - 1 AUUUCAUGCUCAUCUGCAUAAAAUCAAUUCCAUUUUGGUAGACAAUUUGAAGAUUGGAAUACAAAUUUCGAAUUAUCAACAUGGGACCCCC----------------------------- ((((.((((......)))).))))...((((((.((((((....((((((((.(((.....))).)))))))))))))).)))))).....----------------------------- ( -21.20) >DroWil_CAF1 294303 119 - 1 AUUUCAUGCUCAUCUGCAUAAAAUCAAUUCCAUUUUGG-AGACAAUUUGAAGAUUGGAAUACAAAUUUCGAAUUAUCAACCUGGGACCAGAAAAAAAAGAGGACAACCAGAAGAAGUAUA (((((((((......)))).............((((((-.((.(((((((((.(((.....))).))))))))).))...(((....)))................)))))))))))... ( -25.40) >DroAna_CAF1 252211 89 - 1 AUUUCAUGCUCAUCUGCAUAAAAUCAAUUCCAUUUUGG-AGACAAUUUGAAGAUUGGAAUACAAAUUUCGAAUUAUCAACAGGGGACCCA------------------------------ ((((.((((......)))).))))....(((.....))-)((.(((((((((.(((.....))).))))))))).))....((....)).------------------------------ ( -19.10) >DroPer_CAF1 262608 91 - 1 AUUUCAUGCUCAUCUGCAUAAAAUCAAUUCCAUUUUGGUAGACAAUUUGAAGAUUGGAAUACAAAUUUCGAAUUAUCAACAUGGGACCCCC----------------------------- ((((.((((......)))).))))...((((((.((((((....((((((((.(((.....))).)))))))))))))).)))))).....----------------------------- ( -21.20) >consensus AUUUCAUGCUCAUCUGCAUAAAAUCAAUUCCAUUUUGG_AGACAAUUUGAAGAUUGGAAUACAAAUUUCGAAUUAUCAACAUGGGACCCCA_____________________________ ((((.((((......)))).))))...((((((.((((.....(((((((((.(((.....))).))))))))).)))).)))))).................................. (-18.18 = -18.40 + 0.22)


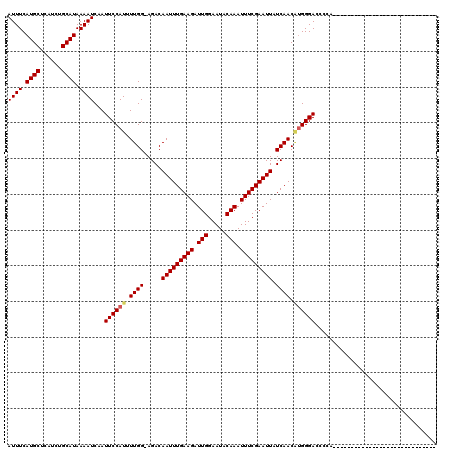
| Location | 24,047,632 – 24,047,751 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.66 |
| Mean single sequence MFE | -21.27 |
| Consensus MFE | -18.63 |
| Energy contribution | -18.13 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.27 |
| SVM RNA-class probability | 0.938948 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24047632 119 - 27905053 ACAUUUAUUUACACCAGCAUGCAAAACUCGCAUUGAGACAAUUUCAUGCUCAUCUGCAUAAAAUCAAUUCCAUUUUGG-AGACAAUUUGAAGAUUGGAAUACAAAUUUCGAAUUAUCAAC .............((((.(((.....(((.....)))...((((.((((......)))).))))......))).))))-.((.(((((((((.(((.....))).))))))))).))... ( -21.30) >DroVir_CAF1 265375 119 - 1 GCAUUUAUUUACUCUAACAUGGUAAAUUCGCAUUGAGACAAUUUCAUGCUCAUCUGCAUAAAAUCAAUUCCAUUUUGG-AGACAAUUUGAAGAUUGGAAUACAAAUUUCGAAUUAUCAAC ...........((((((.((((.......(((.((((.((......))))))..)))............)))).))))-))..(((((((((.(((.....))).)))))))))...... ( -23.51) >DroPse_CAF1 261215 120 - 1 ACAUUUAUUUACACUAACAUGCAAAACUCGCAUUGAGACAAUUUCAUGCUCAUCUGCAUAAAAUCAAUUCCAUUUUGGUAGACAAUUUGAAGAUUGGAAUACAAAUUUCGAAUUAUCAAC ............(((((.(((.....(((.....)))...((((.((((......)))).))))......))).))))).((.(((((((((.(((.....))).))))))))).))... ( -20.40) >DroGri_CAF1 222658 119 - 1 GCAUUUAUUUACACUAACAUGGUAAAUUCGCAUUGAGACAAUUUCAUGCUCAUCUGCAUAAAAUCAAUUCCAUUUUGG-AGACAAUUUGAAGAUUGGAAUACAAAUUUCGAAUUAUCAAC ......((((((.(......)))))))..(((.((((.((......))))))..)))...........(((.....))-)((.(((((((((.(((.....))).))))))))).))... ( -22.40) >DroWil_CAF1 294343 119 - 1 ACAUUUAUUUACACUAACAUGCAAAACUCGCUUUGAGACAAUUUCAUGCUCAUCUGCAUAAAAUCAAUUCCAUUUUGG-AGACAAUUUGAAGAUUGGAAUACAAAUUUCGAAUUAUCAAC .............................((..((((.((......))))))...))...........(((.....))-)((.(((((((((.(((.....))).))))))))).))... ( -19.60) >DroPer_CAF1 262619 120 - 1 ACAUUUAUUUACACUAACAUGCAAAACUCGCAUUGAGACAAUUUCAUGCUCAUCUGCAUAAAAUCAAUUCCAUUUUGGUAGACAAUUUGAAGAUUGGAAUACAAAUUUCGAAUUAUCAAC ............(((((.(((.....(((.....)))...((((.((((......)))).))))......))).))))).((.(((((((((.(((.....))).))))))))).))... ( -20.40) >consensus ACAUUUAUUUACACUAACAUGCAAAACUCGCAUUGAGACAAUUUCAUGCUCAUCUGCAUAAAAUCAAUUCCAUUUUGG_AGACAAUUUGAAGAUUGGAAUACAAAUUUCGAAUUAUCAAC .............((((.(((.....(((.....)))...((((.((((......)))).))))......))).))))..((.(((((((((.(((.....))).))))))))).))... (-18.63 = -18.13 + -0.50)

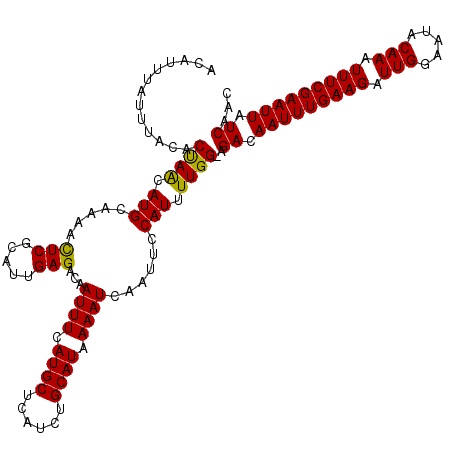

| Location | 24,047,672 – 24,047,782 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.70 |
| Mean single sequence MFE | -26.48 |
| Consensus MFE | -14.04 |
| Energy contribution | -14.65 |
| Covariance contribution | 0.61 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.53 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24047672 110 + 27905053 U-CCAAAAUGGAAUUGAUUUUAUGCAGAUGAGCAUGAAAUUGUCUCAAUGCGAGUUUUGCAUGCUGGUGUAAAUAAAUGUAAAGAUGUCCUGGC---------CGAGGGAACAAGGACAA (-((.....)))...((((((((((......))))))))))((((...((((...(((((((....)))))))....)))).))))(((((..(---------(...))....))))).. ( -26.80) >DroVir_CAF1 265415 97 + 1 U-CCAAAAUGGAAUUGAUUUUAUGCAGAUGAGCAUGAAAUUGUCUCAAUGCGAAUUUACCAUGUUAGAGUAAAUAAAUGCUGCACAGUC---------------G--G-----UGAACUA (-((.....)))(((((((...(((((.((((((......)).))))......(((((((......).)))))).....))))).))))---------------)--)-----)...... ( -19.80) >DroPse_CAF1 261255 120 + 1 UACCAAAAUGGAAUUGAUUUUAUGCAGAUGAGCAUGAAAUUGUCUCAAUGCGAGUUUUGCAUGUUAGUGUAAAUAAAUGUCACGAUGCGAUGGCGUCACGAGGCACAGGGGCAAGGACGA ..((...........((((((((((......))))))))))(((((..(((....(((((((....)))))))..........(((((....))))).....)))..)))))..)).... ( -32.80) >DroGri_CAF1 222698 99 + 1 U-CCAAAAUGGAAUUGAUUUUAUGCAGAUGAGCAUGAAAUUGUCUCAAUGCGAAUUUACCAUGUUAGUGUAAAUAAAUGCUGCACAGUC---------------GAGG-----CGAACUA (-((.....)))...((((((((((......))))))))))(((((.(((.(......))))....(((((.........)))))....---------------))))-----)...... ( -22.10) >DroWil_CAF1 294383 99 + 1 U-CCAAAAUGGAAUUGAUUUUAUGCAGAUGAGCAUGAAAUUGUCUCAAAGCGAGUUUUGCAUGUUAGUGUAAAUAAAUGUAACGACGG----UU---------CAAGAG-ACAA------ (-((.....)))...((((((((((......))))))))))(((((.........(((((((....)))))))....((.(((....)----))---------)).)))-))..------ ( -24.60) >DroPer_CAF1 262659 120 + 1 UACCAAAAUGGAAUUGAUUUUAUGCAGAUGAGCAUGAAAUUGUCUCAAUGCGAGUUUUGCAUGUUAGUGUAAAUAAAUGUCACGAUGCGAUGGCGUCACGAGGCACAGGGGCAAGGACGA ..((...........((((((((((......))))))))))(((((..(((....(((((((....)))))))..........(((((....))))).....)))..)))))..)).... ( -32.80) >consensus U_CCAAAAUGGAAUUGAUUUUAUGCAGAUGAGCAUGAAAUUGUCUCAAUGCGAGUUUUGCAUGUUAGUGUAAAUAAAUGUAACGAUGUC___GC_________CAAGGG__CAAGGACGA ..........((..(((((((((((......)))))))))))..))...(((...(((((((....)))))))....)))........................................ (-14.04 = -14.65 + 0.61)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:23:43 2006