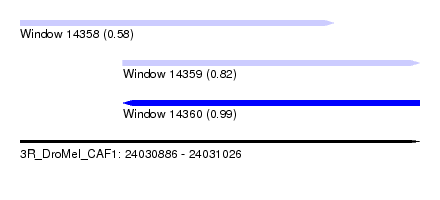
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 24,030,886 – 24,031,026 |
| Length | 140 |
| Max. P | 0.988496 |

| Location | 24,030,886 – 24,030,996 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 72.29 |
| Mean single sequence MFE | -18.44 |
| Consensus MFE | -5.04 |
| Energy contribution | -4.72 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.27 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.575759 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24030886 110 + 27905053 GCUUCAAAUUAAAAAGCAAUAUAACAAAUUCAAUACAAAAGUUUGGAAAAUGAGGACCAGAAAGUCGAAUUUAUUCCUAUAAUCCUACUUUCCAGCUCUGGCC-----AAACUUU ((((.........))))....................(((((((((.....(((.....((((((.(...((((....))))..).))))))...)))...))-----))))))) ( -18.30) >DroSec_CAF1 226160 110 + 1 GCUUUAAAUUAAAAAGCAAUAUAACAUAAGCAAUACAAAAGUUUGGAUAUUGAGGACCGGAAAGCCGAAUUUAUUCCUAUAAUCCUACUUUCCAGCUUUGACA-----AAACUUU (((((.......)))))....................((((((.(((.....((((.(((....)))...((((....))))))))....)))))))))....-----....... ( -17.70) >DroSim_CAF1 223693 110 + 1 GCUUUAAAUUAAAAAGCAAUAUAACAUAAGCAAUACAAAAGUUUGGAUAUUGAGGACCGGAAAGCCGAAUUUAUUCCUAUAAUCCUACUUUCCAGCUUUGACC-----AAACUUU (((((.......)))))....................(((((((((...(..(((.(.((((((......((((....)))).....)))))).))))..)))-----))))))) ( -22.80) >DroEre_CAF1 237584 92 + 1 GCUUUCAACUUA--------------UAAGCAAUGCAAAAGUUUGG---------CCCAGAAUGUCGUAUUUACUGCUGUAAUCCUUCUUUCCACAUUUGGGCAUUUGGAACUUU ((((........--------------.))))......(((((((.(---------((((((.(((.(...((((....)))).........).)))))))))).....))))))) ( -17.30) >DroYak_CAF1 236203 87 + 1 GCUUUAAAUUUA--------------UAAGCAAUACAAAAGUUUGG---------UUCAGAAAGUCGUUUUUCUUUCUGUAAUCCUUCUUUCACCAUUUGGCA-----AAACUUU ((((........--------------.))))......(((((((((---------(((((((((........))))))).)))).........((....))..-----))))))) ( -16.10) >consensus GCUUUAAAUUAAAAAGCAAUAUAACAUAAGCAAUACAAAAGUUUGGA_A_UGAGGACCAGAAAGUCGAAUUUAUUCCUAUAAUCCUACUUUCCAGCUUUGGCC_____AAACUUU .....................................(((((((((..........)).(((((......((((....)))).....)))))................))))))) ( -5.04 = -4.72 + -0.32)


| Location | 24,030,922 – 24,031,026 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 78.69 |
| Mean single sequence MFE | -19.46 |
| Consensus MFE | -8.30 |
| Energy contribution | -8.14 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.43 |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.822981 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24030922 104 + 27905053 AAAAGUUUGGAAAAUGAGGACCAGAAAGUCGAAUUUAUUCCUAUAAUCCUACUUUCCAGCUCUGGCC-----AAACUUUUUUUUUCCUGUGUACCCACACAAAACUUAU ((((((((((.....(((.....((((((.(...((((....))))..).))))))...)))...))-----)))))))).......(((((....)))))........ ( -20.80) >DroSec_CAF1 226196 102 + 1 AAAAGUUUGGAUAUUGAGGACCGGAAAGCCGAAUUUAUUCCUAUAAUCCUACUUUCCAGCUUUGACA-----AAACUUU--UUUUCCCGUGCACCCACACAAAACUUAU ((((((((.....(..(((.(.((((((......((((....)))).....)))))).))))..)..-----)))))))--)......(((......)))......... ( -16.20) >DroSim_CAF1 223729 103 + 1 AAAAGUUUGGAUAUUGAGGACCGGAAAGCCGAAUUUAUUCCUAUAAUCCUACUUUCCAGCUUUGACC-----AAACUUUU-UUUUCCCGUGUACCCACACAAAACUUAU ((((((((((...(..(((.(.((((((......((((....)))).....)))))).))))..)))-----))))))))-.......((((....))))......... ( -23.10) >DroEre_CAF1 237606 98 + 1 AAAAGUUUGG---------CCCAGAAUGUCGUAUUUACUGCUGUAAUCCUUCUUUCCACAUUUGGGCAUUUGGAACUUU--UUUUCCUGUGUAUCCACGCAAAACUUAU ..((((((.(---------((((((.(((.(...((((....)))).........).))))))))))....((((....--..))))(((((....))))))))))).. ( -20.90) >DroYak_CAF1 236225 93 + 1 AAAAGUUUGG---------UUCAGAAAGUCGUUUUUCUUUCUGUAAUCCUUCUUUCACCAUUUGGCA-----AAACUUU--UUUUCCCGUGCAUCUACACAAAACUUAU ((((((((((---------(((((((((........))))))).)))).........((....))..-----)))))))--)......(((......)))......... ( -16.30) >consensus AAAAGUUUGGA_A_UGAGGACCAGAAAGUCGAAUUUAUUCCUAUAAUCCUACUUUCCAGCUUUGGCC_____AAACUUU__UUUUCCCGUGUACCCACACAAAACUUAU ..((((((...............(((((......((((....)))).....)))))................................(((......))).)))))).. ( -8.30 = -8.14 + -0.16)



| Location | 24,030,922 – 24,031,026 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 78.69 |
| Mean single sequence MFE | -25.66 |
| Consensus MFE | -15.80 |
| Energy contribution | -16.64 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.56 |
| Structure conservation index | 0.62 |
| SVM decision value | 2.12 |
| SVM RNA-class probability | 0.988496 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24030922 104 - 27905053 AUAAGUUUUGUGUGGGUACACAGGAAAAAAAAAGUUU-----GGCCAGAGCUGGAAAGUAGGAUUAUAGGAAUAAAUUCGACUUUCUGGUCCUCAUUUUCCAAACUUUU .....((((.((((....)))).))))..((((((((-----((..((..(..((((((.(((((.........))))).))))))..)..))......)))))))))) ( -30.80) >DroSec_CAF1 226196 102 - 1 AUAAGUUUUGUGUGGGUGCACGGGAAAA--AAAGUUU-----UGUCAAAGCUGGAAAGUAGGAUUAUAGGAAUAAAUUCGGCUUUCCGGUCCUCAAUAUCCAAACUUUU .....((((((((....))))))))..(--(((((((-----.(.....((((((((((.(((((.........))))).)))))))))).........).)))))))) ( -24.64) >DroSim_CAF1 223729 103 - 1 AUAAGUUUUGUGUGGGUACACGGGAAAA-AAAAGUUU-----GGUCAAAGCUGGAAAGUAGGAUUAUAGGAAUAAAUUCGGCUUUCCGGUCCUCAAUAUCCAAACUUUU .....((((.((((....)))).)))).-((((((((-----((.....((((((((((.(((((.........))))).)))))))))).........)))))))))) ( -31.54) >DroEre_CAF1 237606 98 - 1 AUAAGUUUUGCGUGGAUACACAGGAAAA--AAAGUUCCAAAUGCCCAAAUGUGGAAAGAAGGAUUACAGCAGUAAAUACGACAUUCUGGG---------CCAAACUUUU ..((((((...(((....))).((((..--....))))....((((((((((.(.........((((....))))...).))))).))))---------).)))))).. ( -19.40) >DroYak_CAF1 236225 93 - 1 AUAAGUUUUGUGUAGAUGCACGGGAAAA--AAAGUUU-----UGCCAAAUGGUGAAAGAAGGAUUACAGAAAGAAAAACGACUUUCUGAA---------CCAAACUUUU .....((((((((....))))))))..(--(((((((-----..((....))..).....((.((.(((((((........)))))))))---------)).))))))) ( -21.90) >consensus AUAAGUUUUGUGUGGGUACACGGGAAAA__AAAGUUU_____GGCCAAAGCUGGAAAGUAGGAUUAUAGGAAUAAAUUCGACUUUCUGGUCCUCA_U_UCCAAACUUUU .....((((((((....)))))))).....(((((((............(((((((((..(((((.........)))))..)))))))))...........))))))). (-15.80 = -16.64 + 0.84)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:23:34 2006