
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
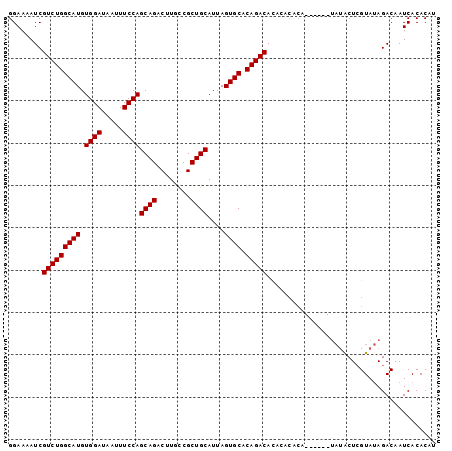
| Location | 24,019,159 – 24,019,328 |
| Length | 169 |
| Max. P | 0.869975 |

| Location | 24,019,159 – 24,019,254 |
|---|---|
| Length | 95 |
| Sequences | 4 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 88.95 |
| Mean single sequence MFE | -23.85 |
| Consensus MFE | -22.10 |
| Energy contribution | -22.10 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.87 |
| SVM RNA-class probability | 0.869975 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24019159 95 + 27905053 GGAAAAUCGUCUGGCAUGUGGAUAAUUUCCAGCAGACUUGCCGCUGCAUUAGUGCACAGACAUACACACA------UAUACUCGUAUAGACAAUCACACAU .((...(((((((((((.((((.....))))((((........))))....)))).))))).........------((((....))))))...))...... ( -22.70) >DroSec_CAF1 214543 87 + 1 GGAAAAUCGUCUGGCAUGUGGAUAAUUUCCAGCAGACUUGUCGCUGCAUUAGUGCACAGACACA--------------CACUCGUAAAGACAAUCACACAU ........(((((((((.((((.....))))((((........))))....)))).)))))...--------------....................... ( -22.10) >DroSim_CAF1 211929 95 + 1 GGAAAAUCGUCUGGCAUGUGGAUAAUUUCCAGCAGACUUGUCGCUGCAUUAGUGCACAGACACAGACACA------UAUACUCGUAUAGACAAUCACACAU ........(((((((((.((((.....))))((((........))))....)))).)))))...((....------((((....)))).....))...... ( -22.80) >DroEre_CAF1 225844 101 + 1 GGAAAAUCGUCUGGCAUGUGGAUAAUUUCCAGCAGACUUGCCGCUGCAUUAGUGCACAGACAGACACACAUACGUGUAUGCUGGUAUAGACAAUCACACAU ........(((((((((.((((.....))))((((........))))....)))).(((.((.((((......)))).)))))...))))).......... ( -27.80) >consensus GGAAAAUCGUCUGGCAUGUGGAUAAUUUCCAGCAGACUUGCCGCUGCAUUAGUGCACAGACACACACACA______UAUACUCGUAUAGACAAUCACACAU ........(((((((((.((((.....))))((((........))))....)))).)))))........................................ (-22.10 = -22.10 + -0.00)


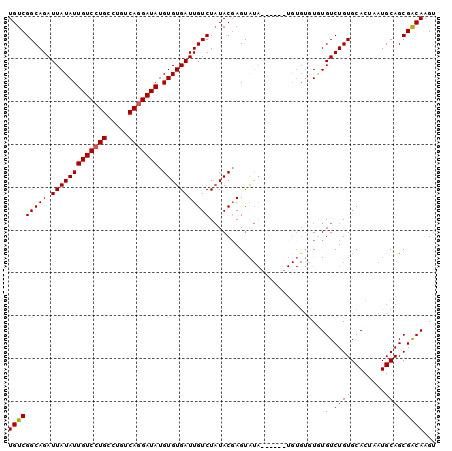
| Location | 24,019,194 – 24,019,288 |
|---|---|
| Length | 94 |
| Sequences | 4 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 87.80 |
| Mean single sequence MFE | -27.33 |
| Consensus MFE | -21.92 |
| Energy contribution | -21.93 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.608594 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24019194 94 - 27905053 UGUCGGCAGAUUAUAUUGUCCUGGCUGUCAGGAUAUGUGUGAUUGUCUAUACGAGUAUA------UGUGUGUAUGUCUGUGCACUAAUGCAGCGGCAAGU (((((((((.(((((((((((((.....))))))).))))))))))).......((((.------.(((..((....))..)))..))))...))))... ( -28.20) >DroSec_CAF1 214578 86 - 1 UGUCGGCAGAUUAUAUUGUCCUGCCUGUCACGAUAUGUGUGAUUGUCUUUACGAGUG--------------UGUGUCUGUGCACUAAUGCAGCGACAAGU (((((((((((......)).))))).(.(((.((((.(((((......))))).)))--------------)))).)..((((....))))..))))... ( -22.60) >DroSim_CAF1 211964 94 - 1 UGUCGGCAGAUUAUAUUGUCCUGCCUGUCAGGAUAUGUGUGAUUGUCUAUACGAGUAUA------UGUGUCUGUGUCUGUGCACUAAUGCAGCGACAAGU (((((((((.(((((((((((((.....))))))).)))))))))))............------(((((..((((....))))..)))))..))))... ( -27.10) >DroEre_CAF1 225879 100 - 1 UGUCGGCAGAUUAUAUUGUCCUGCCUGUCAGGAUAUGUGUGAUUGUCUAUACCAGCAUACACGUAUGUGUGUCUGUCUGUGCACUAAUGCAGCGGCAAGU (((((((((((((((((((((((.....))))))).))))))..........(((.((((((....))))))))))))))(((....)))..)))))... ( -31.40) >consensus UGUCGGCAGAUUAUAUUGUCCUGCCUGUCAGGAUAUGUGUGAUUGUCUAUACGAGUAUA______UGUGUGUGUGUCUGUGCACUAAUGCAGCGACAAGU (((((((((.(((((((((((((.....))))))).)))))))))))...........................(.((((........)))))))))... (-21.92 = -21.93 + 0.00)


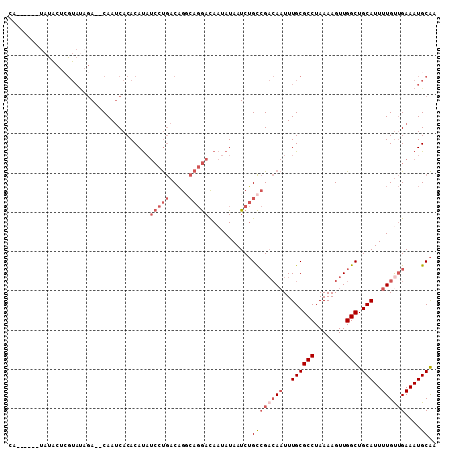
| Location | 24,019,227 – 24,019,328 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 78.71 |
| Mean single sequence MFE | -23.00 |
| Consensus MFE | -13.19 |
| Energy contribution | -15.28 |
| Covariance contribution | 2.09 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.670098 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24019227 101 + 27905053 CA------UAUACUCGUAUAGA--CAAUCACACAUAUCCUGACAGCCAGGACAAUAUAAUCUGCCGACAAUUUGCGCCUAAAAGUUGGCUGCAUUUUGUUGAAAUGCAA ..------.......((((...--............(((((.....))))).............((((((..((((((........))).)))..))))))..)))).. ( -24.50) >DroVir_CAF1 235321 84 + 1 ------------------CAUACGCAAACACACAC----CAACUGCA--AAUAUCAUAAUCAGUGGCAAUUUUGCGCCUAAAAGUUGGCUGCAUUUUGU-GAAAUGCGC ------------------....((((..((((..(----((.(((..--...........))))))......((((((........))).)))...)))-)...)))). ( -17.92) >DroSec_CAF1 214607 97 + 1 ----------CACUCGUAAAGA--CAAUCACACAUAUCGUGACAGGCAGGACAAUAUAAUCUGCCGACAAUUUGCGCCUAAAAGUUGGCUGCAUUUUGUUGAAAUGCAA ----------.....(((....--...((((.......))))..((((((.........))))))(((((..((((((........))).)))..)))))....))).. ( -25.00) >DroSim_CAF1 211997 101 + 1 CA------UAUACUCGUAUAGA--CAAUCACACAUAUCCUGACAGGCAGGACAAUAUAAUCUGCCGACAAUUUGCGCCUAAAAGUUGGCUGCAUUUUGUUGAAAUGCAA ..------.......((((...--............(((((.....))))).............((((((..((((((........))).)))..))))))..)))).. ( -24.30) >DroEre_CAF1 225912 107 + 1 CAUACGUGUAUGCUGGUAUAGA--CAAUCACACAUAUCCUGACAGGCAGGACAAUAUAAUCUGCCGACAAUUUGCGCCUAAAAGUUGGCUGCAUUUUGCUGAAAUGCAA .....((.((((....)))).)--).......(((.((......((((((.........))))))(.(((..((((((........))).)))..)))).)).)))... ( -23.30) >consensus CA______UAUACUCGUAUAGA__CAAUCACACAUAUCCUGACAGGCAGGACAAUAUAAUCUGCCGACAAUUUGCGCCUAAAAGUUGGCUGCAUUUUGUUGAAAUGCAA ....................................(((((.....)))))...........((((((((..((((((........))).)))..))))))....)).. (-13.19 = -15.28 + 2.09)


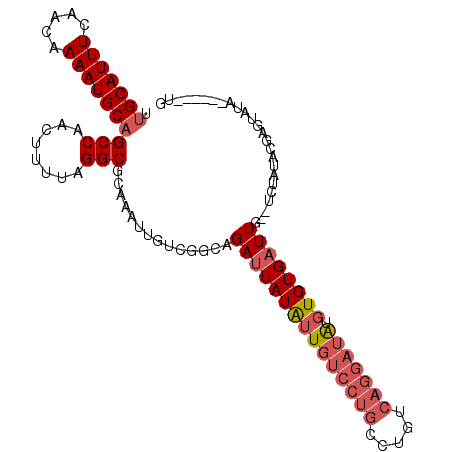
| Location | 24,019,227 – 24,019,328 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 78.71 |
| Mean single sequence MFE | -25.98 |
| Consensus MFE | -16.29 |
| Energy contribution | -17.82 |
| Covariance contribution | 1.53 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.63 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.510923 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24019227 101 - 27905053 UUGCAUUUCAACAAAAUGCAGCCAACUUUUAGGCGCAAAUUGUCGGCAGAUUAUAUUGUCCUGGCUGUCAGGAUAUGUGUGAUUG--UCUAUACGAGUAUA------UG .(((((((.....)))))))(((........))).....((((.(((((.(((((((((((((.....))))))).)))))))))--))...)))).....------.. ( -27.30) >DroVir_CAF1 235321 84 - 1 GCGCAUUUC-ACAAAAUGCAGCCAACUUUUAGGCGCAAAAUUGCCACUGAUUAUGAUAUU--UGCAGUUG----GUGUGUGUUUGCGUAUG------------------ (((((...(-(((...(((.(((........)))))).....(((((((...........--..))).))----)).))))..)))))...------------------ ( -24.02) >DroSec_CAF1 214607 97 - 1 UUGCAUUUCAACAAAAUGCAGCCAACUUUUAGGCGCAAAUUGUCGGCAGAUUAUAUUGUCCUGCCUGUCACGAUAUGUGUGAUUG--UCUUUACGAGUG---------- .(((((((.....)))))))(((........)))(((.(((((.(((((((......)).)))))....))))).)))((((...--...)))).....---------- ( -23.70) >DroSim_CAF1 211997 101 - 1 UUGCAUUUCAACAAAAUGCAGCCAACUUUUAGGCGCAAAUUGUCGGCAGAUUAUAUUGUCCUGCCUGUCAGGAUAUGUGUGAUUG--UCUAUACGAGUAUA------UG .(((((((.....)))))))(((........))).....((((.(((((.(((((((((((((.....))))))).)))))))))--))...)))).....------.. ( -26.60) >DroEre_CAF1 225912 107 - 1 UUGCAUUUCAGCAAAAUGCAGCCAACUUUUAGGCGCAAAUUGUCGGCAGAUUAUAUUGUCCUGCCUGUCAGGAUAUGUGUGAUUG--UCUAUACCAGCAUACACGUAUG .(((......((((..(((.(((........))))))..)))).(((((.(((((((((((((.....))))))).)))))))))--)).......))).......... ( -28.30) >consensus UUGCAUUUCAACAAAAUGCAGCCAACUUUUAGGCGCAAAUUGUCGGCAGAUUAUAUUGUCCUGCCUGUCAGGAUAUGUGUGAUUG__UCUAUACGAGUAUA______UG .(((((((.....)))))))(((........)))..............(((((((((((((((.....))))))).))))))))......................... (-16.29 = -17.82 + 1.53)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:23:31 2006