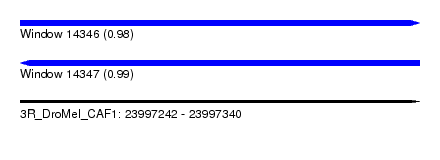
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 23,997,242 – 23,997,340 |
| Length | 98 |
| Max. P | 0.990646 |

| Location | 23,997,242 – 23,997,340 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 72.21 |
| Mean single sequence MFE | -28.78 |
| Consensus MFE | -13.52 |
| Energy contribution | -13.55 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.47 |
| SVM decision value | 1.81 |
| SVM RNA-class probability | 0.978461 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
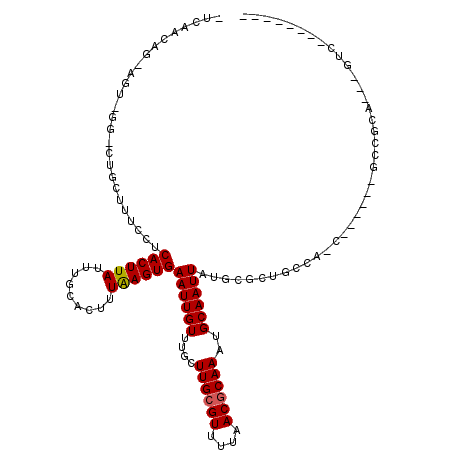
>3R_DroMel_CAF1 23997242 98 + 27905053 AUGGGCAG-AGU-GG-UUUCUCUCCUCACUUAUUUGCACUUUAAGUGAAUUGUUUGCUUGCGUUUUAACGCAAAUGCAAUUAUGCUCUG-----------GCCGGAUUUGUC-------- .(((.(((-(((-..-.........(((((((.........))))))).....((((((((((....))))))..))))....))))))-----------.)))........-------- ( -29.60) >DroVir_CAF1 219784 99 + 1 -UCAGCUG-ACU-GA-CUGCUUUAUGCACUUAUUUACACUUUAAGUGAAUUGUUUGCUUGCGUUUUAACGCAAAUGCAAUUAUGCACAGGCAAUA------UCGCA---UUC-------- -((((...-.))-))-.(((......((((((.........)))))).((((((((.((((((....)))))).((((....)))))))))))).------..)))---...-------- ( -24.60) >DroPse_CAF1 214358 118 + 1 AGAAACAG-AGCUGC-UGGAUUUCCUCACUUAUUUGCACUUUAAGUGAAUUGUUUGCUUGCGUUUUAACGCAAAUGCAAUUAUGCGCUGCCAGCGUCUGGGCCGGGUCCGGCUCGGGACG ........-....((-(((......(((((((.........))))))).......((((((((....))))))..(((....)))))..)))))((((((((((....)))))).)))). ( -41.20) >DroGri_CAF1 175027 96 + 1 -UCAA----ACU-GG-CAGCUUUCUGCACUUAUUUACACUUUAAGUGAAUUGUUUGCUUGCGUUUUAACACAAAUGCAAUUAUGCGCUGCCAACA------ACGCU---GUC-------- -....----..(-((-((((......((((((.........))))))....((.((.((((((((......)))))))).)).)))))))))...------.....---...-------- ( -25.80) >DroWil_CAF1 248080 100 + 1 --UUGCAC-AGUUGGUUGCCUUUCCUCACUUAUUUGCAGUUUAAGUGAAUUGUUUGCUUGCGUUUUAACGCAAAUGCAAUUAUUCACUGCCAGC------GCCACA---AUC-------- --......-.(((((((((................))).....(((((((...((((((((((....))))))..))))..)))))))))))))------......---...-------- ( -25.69) >DroAna_CAF1 210169 99 + 1 AUGGAAGGAAGC-GG-CUGCUUUCCUCACUUAUUUGCACUCUGAGUGAAUUGUUUGCUUGCGUUUUAACGCAAAUGCAAUUAUGCGCUG-----------CCCGGAUUUGCC-------- ......((..((-((-(.((.....(((((((.........))))))).....((((((((((....))))))..))))....))))))-----------))).........-------- ( -25.80) >consensus _UCAACAG_AGU_GG_CUGCUUUCCUCACUUAUUUGCACUUUAAGUGAAUUGUUUGCUUGCGUUUUAACGCAAAUGCAAUUAUGCGCUGCCA_C______GCCGCA___GUC________ ..........................((((((.........))))))((((((....((((((....))))))..))))))....................................... (-13.52 = -13.55 + 0.03)

| Location | 23,997,242 – 23,997,340 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 72.21 |
| Mean single sequence MFE | -27.71 |
| Consensus MFE | -13.78 |
| Energy contribution | -13.92 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.42 |
| Structure conservation index | 0.50 |
| SVM decision value | 2.22 |
| SVM RNA-class probability | 0.990646 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
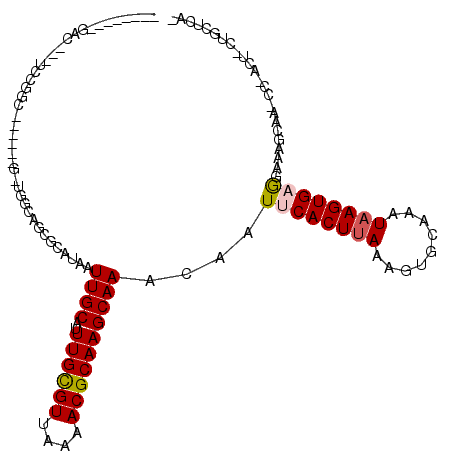
>3R_DroMel_CAF1 23997242 98 - 27905053 --------GACAAAUCCGGC-----------CAGAGCAUAAUUGCAUUUGCGUUAAAACGCAAGCAAACAAUUCACUUAAAGUGCAAAUAAGUGAGGAGAGAAA-CC-ACU-CUGCCCAU --------.........((.-----------(((((.....((((..((((((....)))))))))).(..((((((((.........))))))))..).....-..-.))-))).)).. ( -25.60) >DroVir_CAF1 219784 99 - 1 --------GAA---UGCGA------UAUUGCCUGUGCAUAAUUGCAUUUGCGUUAAAACGCAAGCAAACAAUUCACUUAAAGUGUAAAUAAGUGCAUAAAGCAG-UC-AGU-CAGCUGA- --------...---.((((------((((((.(((((((..((((..((((((....))))))))))......(((.....))).......)))))))..))))-).-.))-).))...- ( -25.40) >DroPse_CAF1 214358 118 - 1 CGUCCCGAGCCGGACCCGGCCCAGACGCUGGCAGCGCAUAAUUGCAUUUGCGUUAAAACGCAAGCAAACAAUUCACUUAAAGUGCAAAUAAGUGAGGAAAUCCA-GCAGCU-CUGUUUCU .(((..(.((((....)))))..)))(((((..(((((....)))..((((((....))))))))......((((((((.........)))))))).....)))-))....-........ ( -37.80) >DroGri_CAF1 175027 96 - 1 --------GAC---AGCGU------UGUUGGCAGCGCAUAAUUGCAUUUGUGUUAAAACGCAAGCAAACAAUUCACUUAAAGUGUAAAUAAGUGCAGAAAGCUG-CC-AGU----UUGA- --------...---.....------..(((((((((((....)))..((((((....))))))(((.......(((.....)))........))).....))))-))-)).----....- ( -26.96) >DroWil_CAF1 248080 100 - 1 --------GAU---UGUGGC------GCUGGCAGUGAAUAAUUGCAUUUGCGUUAAAACGCAAGCAAACAAUUCACUUAAACUGCAAAUAAGUGAGGAAAGGCAACCAACU-GUGCAA-- --------...---....((------((.((((((.....)))))..((((((....))))))((......((((((((.........)))))))).....))......).-))))..-- ( -25.80) >DroAna_CAF1 210169 99 - 1 --------GGCAAAUCCGGG-----------CAGCGCAUAAUUGCAUUUGCGUUAAAACGCAAGCAAACAAUUCACUCAGAGUGCAAAUAAGUGAGGAAAGCAG-CC-GCUUCCUUCCAU --------.((........)-----------).((((....((((..((((((....)))))))))).....((.....))))))......(..(((((.((..-..-)))))))..).. ( -24.70) >consensus ________GAC___UCCGGC______G_UGGCAGCGCAUAAUUGCAUUUGCGUUAAAACGCAAGCAAACAAUUCACUUAAAGUGCAAAUAAGUGAGGAAAGCAA_CC_ACU_CUGCUCA_ .........................................((((..((((((....))))))))))....((((((((.........))))))))........................ (-13.78 = -13.92 + 0.14)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:23:23 2006