
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 23,993,672 – 23,993,772 |
| Length | 100 |
| Max. P | 0.847369 |

| Location | 23,993,672 – 23,993,772 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 78.65 |
| Mean single sequence MFE | -12.88 |
| Consensus MFE | -9.58 |
| Energy contribution | -9.92 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -0.92 |
| Structure conservation index | 0.74 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.509966 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
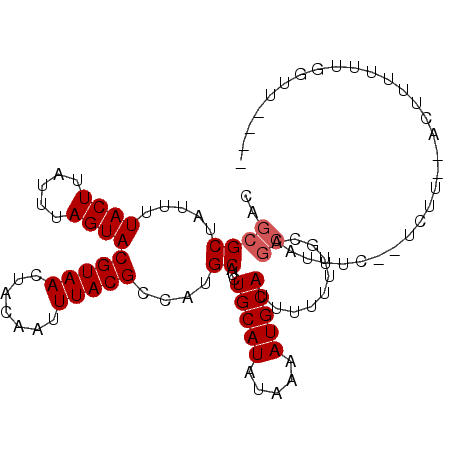
>3R_DroMel_CAF1 23993672 100 + 27905053 CAGCGCUAUUUUACUUAUUUAGUACGUAACUACAAUUUACGCCAUGCAUCUGCAUAUAAAAUGCAUUUUUUUGCAGCAUUUC--UUUUUCACUUUUUUGGUU---- ....(((((((((...........(((((.......)))))..((((....)))).))))))(((......)))))).....--..................---- ( -12.80) >DroVir_CAF1 215427 82 + 1 CAGCGCAAUUUUACUUAUUUAGUACGUAACUGCAAUUUACGCCAUGCAUCUGCAUAUAAAAUGCAUUUUUUUGCAGCCUUUU------------------------ ..((((((...((((.....))))......((((.((((....((((....)))).)))).)))).....)))).)).....------------------------ ( -13.40) >DroPse_CAF1 209812 102 + 1 CAGCGCUAUUUUACUUAUUUAGUACGUAACUACAAUUUACGCCAUGCAUCUGCAUAUAAAAUGCAUUU-UUUGCAGCAUUUC--UCUU-GACUUUUUGGAUACCCU ..((((.....((((.....)))).((((.......))))))..((((..(((((.....)))))...-..)))))).....--....-........((....)). ( -13.50) >DroWil_CAF1 243978 78 + 1 CAGUGCUAUUUUACUUAUUUAGUACGUAACUACAAUUUACGCCAUGCAUCUGCAUAUAAAAUGCAUUUUUUU------------------------UUUGCU---- .(((((.((((((...........(((((.......)))))..((((....)))).))))))))))).....------------------------......---- ( -11.80) >DroMoj_CAF1 221388 100 + 1 CAGCGCGAUUUUACUUAUUUAGUACGUAACUGCAAUUUACGCCAUGCAUCUGCAUACAAAAUGCAUUUUUU--CAGCUUUAUUUUCUUUUCUUCAUUUUGUU---- .((((((....((((.....))))(((((.......)))))...)))...(((((.....)))))......--..)))........................---- ( -12.30) >DroPer_CAF1 211668 102 + 1 CAGCGCUAUUUUACUUAUUUAGUACGUAACUACAAUUUACGCCAUGCAUCUGCAUAUAAAAUGCAUUU-UUUGCAGCAUUUC--UCUU-GACUUUUUGGAUACCCU ..((((.....((((.....)))).((((.......))))))..((((..(((((.....)))))...-..)))))).....--....-........((....)). ( -13.50) >consensus CAGCGCUAUUUUACUUAUUUAGUACGUAACUACAAUUUACGCCAUGCAUCUGCAUAUAAAAUGCAUUUUUUUGCAGCAUUUC__UCUU__ACUUUUUUGGUU____ ..((((.....((((.....))))(((((.......)))))....))...(((((.....)))))..........))............................. ( -9.58 = -9.92 + 0.33)

| Location | 23,993,672 – 23,993,772 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 78.65 |
| Mean single sequence MFE | -16.55 |
| Consensus MFE | -13.23 |
| Energy contribution | -12.95 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.77 |
| SVM RNA-class probability | 0.847369 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
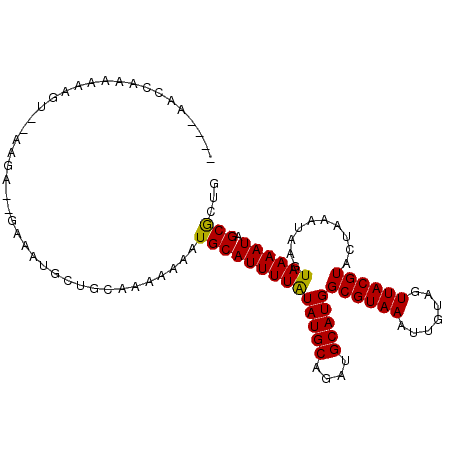
>3R_DroMel_CAF1 23993672 100 - 27905053 ----AACCAAAAAAGUGAAAAA--GAAAUGCUGCAAAAAAAUGCAUUUUAUAUGCAGAUGCAUGGCGUAAAUUGUAGUUACGUACUAAAUAAGUAAAAUAGCGCUG ----.........((((.....--(((((((...........))))))).(((((....)))))((((((.......))))))..................)))). ( -17.10) >DroVir_CAF1 215427 82 - 1 ------------------------AAAAGGCUGCAAAAAAAUGCAUUUUAUAUGCAGAUGCAUGGCGUAAAUUGCAGUUACGUACUAAAUAAGUAAAAUUGCGCUG ------------------------....(((.((......((((((((.......)))))))).)).......((((((...((((.....)))).))))))))). ( -18.30) >DroPse_CAF1 209812 102 - 1 AGGGUAUCCAAAAAGUC-AAGA--GAAAUGCUGCAAA-AAAUGCAUUUUAUAUGCAGAUGCAUGGCGUAAAUUGUAGUUACGUACUAAAUAAGUAAAAUAGCGCUG ..(((.........(((-....--(((((((......-....)))))))..((((....)))))))..........((((..((((.....))))...))))))). ( -17.20) >DroWil_CAF1 243978 78 - 1 ----AGCAAA------------------------AAAAAAAUGCAUUUUAUAUGCAGAUGCAUGGCGUAAAUUGUAGUUACGUACUAAAUAAGUAAAAUAGCACUG ----......------------------------.......((((((((((((((....)))).((((((.......)))))).........))))))).)))... ( -14.00) >DroMoj_CAF1 221388 100 - 1 ----AACAAAAUGAAGAAAAGAAAAUAAAGCUG--AAAAAAUGCAUUUUGUAUGCAGAUGCAUGGCGUAAAUUGCAGUUACGUACUAAAUAAGUAAAAUCGCGCUG ----........................(((((--(....((((((((.(....))))))))).((((((.......))))))...............))).))). ( -15.50) >DroPer_CAF1 211668 102 - 1 AGGGUAUCCAAAAAGUC-AAGA--GAAAUGCUGCAAA-AAAUGCAUUUUAUAUGCAGAUGCAUGGCGUAAAUUGUAGUUACGUACUAAAUAAGUAAAAUAGCGCUG ..(((.........(((-....--(((((((......-....)))))))..((((....)))))))..........((((..((((.....))))...))))))). ( -17.20) >consensus ____AACCAAAAAAGU__AAGA__GAAAUGCUGCAAAAAAAUGCAUUUUAUAUGCAGAUGCAUGGCGUAAAUUGUAGUUACGUACUAAAUAAGUAAAAUAGCGCUG .........................................((((((((((((((....)))))((((((.......))))))..........)))))).)))... (-13.23 = -12.95 + -0.28)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:23:18 2006