
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 23,980,759 – 23,980,880 |
| Length | 121 |
| Max. P | 0.920153 |

| Location | 23,980,759 – 23,980,859 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.73 |
| Mean single sequence MFE | -18.87 |
| Consensus MFE | -15.24 |
| Energy contribution | -15.35 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.88 |
| SVM RNA-class probability | 0.873915 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
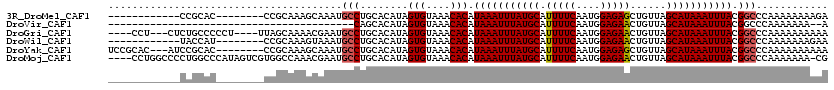
>3R_DroMel_CAF1 23980759 100 + 27905053 ------------CCGCAC--------CCGCAAAGCAAAUGCCUGCACAUAGUGUAAACACAUAAAUUUAUGCAUUUUCAAUGGAGAGCUGUUAGCAUAAAUUUACGGCCCAAAAAAAAGA ------------(((...--------..(((..((....)).))).....(((....))).(((((((((((.(((((....)))))......))))))))))))))............. ( -18.00) >DroVir_CAF1 201997 77 + 1 -----------------------------------------CAGCACAUAGUGUAAACACAUAAAUUUAUGCAUUUUCAAUGGAGAACUGUUAGCAUAAAUUUACGGCCCAAAAAAA--A -----------------------------------------..((.....(((....))).(((((((((((.(((((....)))))......)))))))))))..)).........--. ( -13.80) >DroGri_CAF1 157450 109 + 1 ----CCU---CUCUGCCCCCU----UUAGCAAAACGAAUGCCUGCACAUAGUGUAAACACAUAAAUUUAUGCAUUUUCAAUGGAGAACUGUUAGCAUAAAUUUACGGCCCAAAAAAAAAA ----...---...(((.....----...)))........((((((((...)))))......(((((((((((.(((((....)))))......))))))))))).)))............ ( -18.00) >DroWil_CAF1 227557 100 + 1 ------------UACCAU--------CCGCAAAGUAAAUGCCUGCACAUAGUGUAAACACAUAAAUUUAUGCAUUUUCAAUGGAGAACUGUUAGCAUAAAUUUACGGCCCAAAAAAAGAA ------------......--------.............((((((((...)))))......(((((((((((.(((((....)))))......))))))))))).)))............ ( -16.70) >DroYak_CAF1 191652 109 + 1 UCCGCAC---AUCCGCAC--------CCGCAAAGCAAAUGCCUGCACAUAGUGUAAACACAUAAAUUUAUGCAUUUUCAAUGGAGAGCUGUUAGCAUAAAUUUACGGCCCAAAAAAAAAA .((((((---....((..--------..))...(((......))).....)))).......(((((((((((.(((((....)))))......))))))))))).))............. ( -19.10) >DroMoj_CAF1 206760 115 + 1 ----CCUGGCCCCUGGCCCAUAGUCGUGGCCAAACGAAUGCCUGCACAUAGUGUAAACACAUAAAUUUAUGCAUUUUCAAUGGAGAACUGUUAGCAUAAAUUUACGGCCCAAAAAAA-CG ----...((((..((((((......).)))))......((..(((((...)))))..))..(((((((((((.(((((....)))))......))))))))))).))))........-.. ( -27.60) >consensus ____________CCGCAC________CCGCAAAGCAAAUGCCUGCACAUAGUGUAAACACAUAAAUUUAUGCAUUUUCAAUGGAGAACUGUUAGCAUAAAUUUACGGCCCAAAAAAAAAA .......................................(((........(((....))).(((((((((((.(((((....)))))......))))))))))).)))............ (-15.24 = -15.35 + 0.11)


| Location | 23,980,759 – 23,980,859 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.73 |
| Mean single sequence MFE | -26.80 |
| Consensus MFE | -17.48 |
| Energy contribution | -18.15 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.65 |
| SVM decision value | 1.13 |
| SVM RNA-class probability | 0.920153 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 23980759 100 - 27905053 UCUUUUUUUUGGGCCGUAAAUUUAUGCUAACAGCUCUCCAUUGAAAAUGCAUAAAUUUAUGUGUUUACACUAUGUGCAGGCAUUUGCUUUGCGG--------GUGCGG------------ .........((((((((((((((((((......(........).....))))))))))))).)))))............((((((((...))))--------))))..------------ ( -24.70) >DroVir_CAF1 201997 77 - 1 U--UUUUUUUGGGCCGUAAAUUUAUGCUAACAGUUCUCCAUUGAAAAUGCAUAAAUUUAUGUGUUUACACUAUGUGCUG----------------------------------------- .--......((((((((((((((((((......(((......)))...))))))))))))).)))))............----------------------------------------- ( -18.00) >DroGri_CAF1 157450 109 - 1 UUUUUUUUUUGGGCCGUAAAUUUAUGCUAACAGUUCUCCAUUGAAAAUGCAUAAAUUUAUGUGUUUACACUAUGUGCAGGCAUUCGUUUUGCUAA----AGGGGGCAGAG---AGG---- ............(((((((((((((((......(((......)))...))))))))))))(((....)))........))).....(((((((..----....)))))))---...---- ( -26.20) >DroWil_CAF1 227557 100 - 1 UUCUUUUUUUGGGCCGUAAAUUUAUGCUAACAGUUCUCCAUUGAAAAUGCAUAAAUUUAUGUGUUUACACUAUGUGCAGGCAUUUACUUUGCGG--------AUGGUA------------ .........((((((((((((((((((......(((......)))...))))))))))))).))))).(((((.((((((.......)))))).--------))))).------------ ( -27.60) >DroYak_CAF1 191652 109 - 1 UUUUUUUUUUGGGCCGUAAAUUUAUGCUAACAGCUCUCCAUUGAAAAUGCAUAAAUUUAUGUGUUUACACUAUGUGCAGGCAUUUGCUUUGCGG--------GUGCGGAU---GUGCGGA .........((((((((((((((((((......(........).....))))))))))))).))))).....((..((.((((((((...))))--------))))...)---)..)).. ( -29.70) >DroMoj_CAF1 206760 115 - 1 CG-UUUUUUUGGGCCGUAAAUUUAUGCUAACAGUUCUCCAUUGAAAAUGCAUAAAUUUAUGUGUUUACACUAUGUGCAGGCAUUCGUUUGGCCACGACUAUGGGCCAGGGGCCAGG---- ..-........((((((((((((((((......(((......)))...))))))))))))((((((.(((...))).))))))...(((((((.(......))))))))))))...---- ( -34.60) >consensus UUUUUUUUUUGGGCCGUAAAUUUAUGCUAACAGUUCUCCAUUGAAAAUGCAUAAAUUUAUGUGUUUACACUAUGUGCAGGCAUUUGCUUUGCGG________GUGCAG____________ ............(((((((((((((((......(((......)))...))))))))))))(((....)))........)))....................................... (-17.48 = -18.15 + 0.67)


| Location | 23,980,779 – 23,980,880 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 83.77 |
| Mean single sequence MFE | -16.47 |
| Consensus MFE | -13.76 |
| Energy contribution | -13.53 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.617763 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
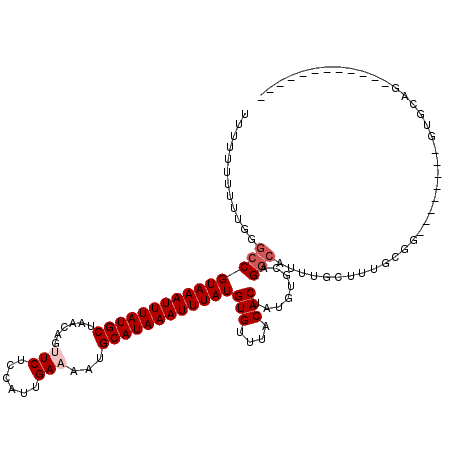
>3R_DroMel_CAF1 23980779 101 + 27905053 CCUGCACAUAGUGUAAACACAUAAAUUUAUGCAUUUUCAAUGGAGAGCUGUUAGCAUAAAUUUACGGCCCAAAAAAAAGA-CGACAAAAAAUGGGGAA---AAUC-- (((((((...)))))......(((((((((((.(((((....)))))......))))))))))).))((((.........-..........))))...---....-- ( -15.91) >DroVir_CAF1 201997 92 + 1 -CAGCACAUAGUGUAAACACAUAAAUUUAUGCAUUUUCAAUGGAGAACUGUUAGCAUAAAUUUACGGCCCAAAAAAA--ACCAACAUAG-CU---GAG---A----- -((((.....(((....))).(((((((((((.(((((....)))))......))))))))))).............--.........)-))---)..---.----- ( -18.30) >DroGri_CAF1 157479 102 + 1 CCUGCACAUAGUGUAAACACAUAAAUUUAUGCAUUUUCAAUGGAGAACUGUUAGCAUAAAUUUACGGCCCAAAAAAAAAACCCACAAAAACA-G-GAA---UAUGGC ((((......(((....))).(((((((((((.(((((....)))))......)))))))))))..........................))-)-)..---...... ( -17.20) >DroWil_CAF1 227577 104 + 1 CCUGCACAUAGUGUAAACACAUAAAUUUAUGCAUUUUCAAUGGAGAACUGUUAGCAUAAAUUUACGGCCCAAAAAAAGAA-AGAAAAAAAAUACGAAAACGAACC-- ...((.....(((....))).(((((((((((.(((((....)))))......)))))))))))..))............-........................-- ( -13.60) >DroMoj_CAF1 206796 94 + 1 CCUGCACAUAGUGUAAACACAUAAAUUUAUGCAUUUUCAAUGGAGAACUGUUAGCAUAAAUUUACGGCCCAAAAAAA-CGCCAACAAAC-CA---GAG---C----- .(((......(((....))).(((((((((((.(((((....)))))......))))))))))).(((.........-.))).......-))---)..---.----- ( -16.70) >DroAna_CAF1 191064 99 + 1 CCUGCACACAGUGUAAACACAUAAAUUUAUGCAUUUUCAAUGGAGAGCUGUUAGCAUAAAUUUACGGCCCAAAAAAAAGC---ACAAAAUUGGGGAAA---AAUC-- (((((((...)))))......(((((((((((.(((((....)))))......))))))))))).))(((((........---......)))))....---....-- ( -17.14) >consensus CCUGCACAUAGUGUAAACACAUAAAUUUAUGCAUUUUCAAUGGAGAACUGUUAGCAUAAAUUUACGGCCCAAAAAAAAAA_CAACAAAAACU_G_GAA___AAUC__ ...((.....(((....))).(((((((((((.(((((....)))))......)))))))))))..))....................................... (-13.76 = -13.53 + -0.22)


| Location | 23,980,779 – 23,980,880 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 83.77 |
| Mean single sequence MFE | -21.16 |
| Consensus MFE | -16.93 |
| Energy contribution | -17.27 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.563474 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
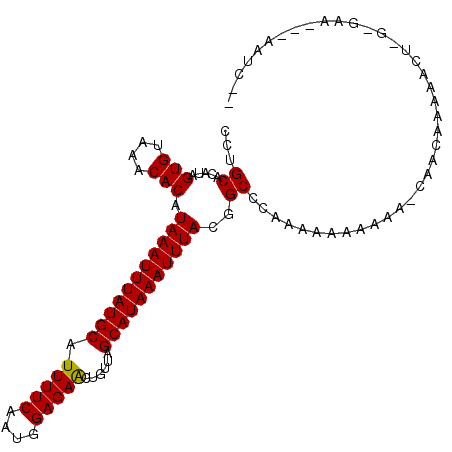
>3R_DroMel_CAF1 23980779 101 - 27905053 --GAUU---UUCCCCAUUUUUUGUCG-UCUUUUUUUUGGGCCGUAAAUUUAUGCUAACAGCUCUCCAUUGAAAAUGCAUAAAUUUAUGUGUUUACACUAUGUGCAGG --....---...((((..........-.........))))((((((((((((((......(........).....))))))))))))(((....)))........)) ( -16.81) >DroVir_CAF1 201997 92 - 1 -----U---CUC---AG-CUAUGUUGGU--UUUUUUUGGGCCGUAAAUUUAUGCUAACAGUUCUCCAUUGAAAAUGCAUAAAUUUAUGUGUUUACACUAUGUGCUG- -----.---..(---((-(((((..(((--((.....)))))((((((((((((......(((......)))...))))))))))))(((....))))))).))))- ( -22.70) >DroGri_CAF1 157479 102 - 1 GCCAUA---UUC-C-UGUUUUUGUGGGUUUUUUUUUUGGGCCGUAAAUUUAUGCUAACAGUUCUCCAUUGAAAAUGCAUAAAUUUAUGUGUUUACACUAUGUGCAGG ......---..(-(-(((....((((((((.......)))))((((((((((((......(((......)))...)))))))))))).......))).....))))) ( -25.70) >DroWil_CAF1 227577 104 - 1 --GGUUCGUUUUCGUAUUUUUUUUCU-UUCUUUUUUUGGGCCGUAAAUUUAUGCUAACAGUUCUCCAUUGAAAAUGCAUAAAUUUAUGUGUUUACACUAUGUGCAGG --((((((..................-.........))))))((((((((((((......(((......)))...))))))))))))(((....))).......... ( -19.13) >DroMoj_CAF1 206796 94 - 1 -----G---CUC---UG-GUUUGUUGGCG-UUUUUUUGGGCCGUAAAUUUAUGCUAACAGUUCUCCAUUGAAAAUGCAUAAAUUUAUGUGUUUACACUAUGUGCAGG -----(---(.(---((-((.....(((.-(......).)))((((((((((((......(((......)))...))))))))))))........)))).).))... ( -22.00) >DroAna_CAF1 191064 99 - 1 --GAUU---UUUCCCCAAUUUUGU---GCUUUUUUUUGGGCCGUAAAUUUAUGCUAACAGCUCUCCAUUGAAAAUGCAUAAAUUUAUGUGUUUACACUGUGUGCAGG --....---...........(((.---.(.......((((((((((((((((((......(........).....))))))))))))).)))))......)..))). ( -20.62) >consensus __GAUU___UUC_C_AGUUUUUGUCG_UCUUUUUUUUGGGCCGUAAAUUUAUGCUAACAGUUCUCCAUUGAAAAUGCAUAAAUUUAUGUGUUUACACUAUGUGCAGG ....................................((((((((((((((((((......(((......)))...))))))))))))).)))))............. (-16.93 = -17.27 + 0.33)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:23:08 2006