| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
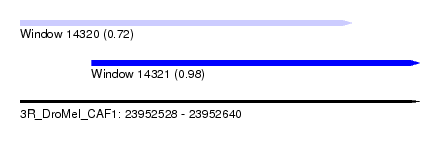
| Location | 23,952,528 – 23,952,640 |
| Length | 112 |
| Max. P | 0.980495 |

| Location | 23,952,528 – 23,952,621 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 86.54 |
| Mean single sequence MFE | -26.73 |
| Consensus MFE | -22.55 |
| Energy contribution | -22.55 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.718367 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
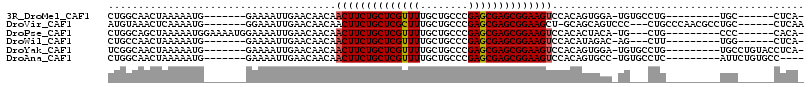
>3R_DroMel_CAF1 23952528 93 + 27905053 UUUUAUCCCCCAUC----------CUUCG------GCUGGCAACUAAAAAUGGAAAAUUGAACAACAACUUCUGCUCGUUUUGCUGCCCGAGCGAGCGGAAGUCCACAG (((((....(((.(----------(...)------).)))....))))).((((...(((.....)))(((((((((((((........)))))))))))))))))... ( -25.20) >DroSec_CAF1 154263 99 + 1 UCUUAUCCCCCAUC----------CUUCGGCAAUGGCUGGCAACUAAAAAUGGAAAAUUGAACAACAACUUCUGCUCGUUUUGCUGCCCGAGCGAGCGGAAGUCCACAG .....(((.((((.----------(....)..))))..(....).......))).............((((((((((((((........))))))))))))))...... ( -26.60) >DroSim_CAF1 155812 99 + 1 UCUUAUCCCCCAUC----------CUUCGGCAAUGGCUGGCAACUAAAAAUGGAAAAUUGAACAACAACUUCUGCUCGUUUUGCUGCCCGAGCGAGCGGAAGUCCACAG .....(((.((((.----------(....)..))))..(....).......))).............((((((((((((((........))))))))))))))...... ( -26.60) >DroEre_CAF1 162306 97 + 1 -UUCAUCC-CCAUC----------CCUCGGCAAUGGCUGGCAACUAAAAAUGGAAAAUUGAACAACAACUUCUGCUCGUUUUGCUGCCCGAGCGAGCGGAAGUCCACAG -((((...-((((.----------....(((....)))(....).....)))).....)))).....((((((((((((((........))))))))))))))...... ( -26.40) >DroYak_CAF1 162344 96 + 1 CUUCCUCC-CCUUC----------CCCCCCUCC--UUCGGCAACUAAAAAUGGAAAAUUGAACAACAACUUCUGCUCGUUUUGCUGCCCGAGCGAGCGGAAGUCCACAG .((((...-.....----------....((...--...))...........))))............((((((((((((((........))))))))))))))...... ( -23.30) >DroAna_CAF1 163032 107 + 1 --CCAUGCCCCAUUCACUUUUUCACUUCGGCAACGACUGGCAACUAAAAAUGGAAAAUUGAACAACAACUUCUGCUCGUUUUGCUGCCCGAGCGAGCGGAAGUCCACAG --..........((((.(((((((..(((....)))..(....)......))))))).)))).....((((((((((((((........))))))))))))))...... ( -32.30) >consensus UUUCAUCCCCCAUC__________CUUCGGCAAUGGCUGGCAACUAAAAAUGGAAAAUUGAACAACAACUUCUGCUCGUUUUGCUGCCCGAGCGAGCGGAAGUCCACAG ......................................(....)......((((...(((.....)))(((((((((((((........)))))))))))))))))... (-22.55 = -22.55 + 0.00)



| Location | 23,952,548 – 23,952,640 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 76.83 |
| Mean single sequence MFE | -29.92 |
| Consensus MFE | -20.14 |
| Energy contribution | -20.17 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.67 |
| SVM decision value | 1.86 |
| SVM RNA-class probability | 0.980495 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 23952548 92 + 27905053 CUGGCAACUAAAAAUG-------GAAAAUUGAACAACAACUUCUGCUCGUUUUGCUGCCCGAGCGAGCGGAAGUCCACAGUGGA-UGUGCCUG---------UGC------CUCA- ..((((.(((....((-------((...(((.....)))(((((((((((((........)))))))))))))))))...))).-..))))..---------...------....- ( -29.50) >DroVir_CAF1 172702 99 + 1 AUGUAAACUCAAAAUG-------GGAAAUUGAACAACAACUUCUGCUCGCUUUGCUGCCCGAGCGAGCGGAAGCU-GCAGCAGUCCC---CUGCCCAACGCCUGC------CUCAA ...............(-------((...(((.....((.(((((((((((((........))))))))))))).)-)..((((....---)))).)))..)))..------..... ( -30.10) >DroPse_CAF1 165597 96 + 1 CUGGCAGCUAAAAAUGGAAAAUGGAAAAUUGAACAACAACUUCUGCUCGUUUUGCUGCCCGAGCGAGCGGAAGUCCACACUACA-UG---CUG---------CCC------CACA- ..((((((.....(((.....((((...(((.....)))(((((((((((((........))))))))))))))))).....))-))---)))---------)).------....- ( -33.40) >DroWil_CAF1 197122 89 + 1 CUGCCAACUAAAAAUG-------GAAAAUUGAACAACAACUUCUGCUCGUUUUGCUGCCCGAGCGAGCGGAAGUCCACAUAGAC-AG---CUU---------UGG------CUCA- ..((((((((....((-------((...(((.....)))(((((((((((((........)))))))))))))))))..)))..-..---..)---------)))------)...- ( -26.70) >DroYak_CAF1 162367 98 + 1 UCGGCAACUAAAAAUG-------GAAAAUUGAACAACAACUUCUGCUCGUUUUGCUGCCCGAGCGAGCGGAAGUCCACAGUGGA-UGUGCCUG---------UGCCUGUACCUCA- ..((((.(((....((-------((...(((.....)))(((((((((((((........)))))))))))))))))...))).-..))))..---------.............- ( -29.80) >DroAna_CAF1 163066 95 + 1 CUGGCAACUAAAAAUG-------GAAAAUUGAACAACAACUUCUGCUCGUUUUGCUGCCCGAGCGAGCGGAAGUCCACAGUGCC-UGUGCCUC---------AUUCUGUGCC---- ..((((.........(-------(..............((((((((((((((........)))))))))))))).(((((...)-))))))..---------......))))---- ( -30.03) >consensus CUGGCAACUAAAAAUG_______GAAAAUUGAACAACAACUUCUGCUCGUUUUGCUGCCCGAGCGAGCGGAAGUCCACAGUGGA_UG___CUG_________UGC______CUCA_ ......................................((((((((((((((........)))))))))))))).......................................... (-20.14 = -20.17 + 0.03)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:22:59 2006