
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 23,921,592 – 23,921,712 |
| Length | 120 |
| Max. P | 0.998384 |

| Location | 23,921,592 – 23,921,693 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 85.98 |
| Mean single sequence MFE | -35.93 |
| Consensus MFE | -33.49 |
| Energy contribution | -33.38 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.09 |
| Mean z-score | -4.36 |
| Structure conservation index | 0.93 |
| SVM decision value | 3.08 |
| SVM RNA-class probability | 0.998384 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 23921592 101 + 27905053 AAGCGGCAGCUGGCCAAAGUCGAAGGAAGUUGGUUGGCUGCCAUGUCAACGGCUAAAUGACACUGGCACUGGGAAUGGAAAUA------GGAAUAGGAAUAGGAAUA ..((((((((..(((((..((....))..)))))..)))))).(((((.........)))))...))................------.................. ( -34.60) >DroSec_CAF1 123236 89 + 1 AAGCGGCAGCUGGCCAAAGUCGAAGGAAGUUGGUUGGCUGCCAUGUCAACGGCUAAAUGACACUGGCACUGGGAAUGGAAAUA------GGA------------AUA ..((((((((..(((((..((....))..)))))..)))))).(((((.........)))))...))................------...------------... ( -34.60) >DroSim_CAF1 124970 95 + 1 AAGCGGCAGCUGGCCAAAGUCGAAGGAAGUUGGUUGGCUGCCAUGUCAACGGCUAAAUGACACUGGCACUGGGAAUGGAAAUA------GGAAUA------GGAAUA ..((((((((..(((((..((....))..)))))..)))))).(((((.........)))))...))................------......------...... ( -34.60) >DroEre_CAF1 126663 107 + 1 AAGCGGCAGCUGGCCAAAGUCGAAGGAAGUUGGUUGGCUGCCAUGUCAACGGCUAAAUGACACUGGCACUGGGAAUGGGAAUGGGAAUGGAAAUAGGAAUAGGAAUA ..((((((((..(((((..((....))..)))))..)))))).(((((.........)))))...))........................................ ( -34.60) >DroYak_CAF1 130461 95 + 1 AAGCGGCAGCUGGCCAAAGUCGAAGGAAGUUGGUUGGCUGCCAUGUCAACGGCUAAAUGACACUGGCACUGGGAAUGGGAAUG------GAAA------UAGGAACA ..((((((((..(((((..((....))..)))))..)))))).(((((.........)))))...))................------....------........ ( -34.60) >DroAna_CAF1 130966 88 + 1 AAGCGGCAGCUGGCCAAAGUCGAAGGAAGUUGGCCGGCUACCAUGUCAACGGCUAAAUGACACUGGCACUGGGAUGGGUCCUG------GG-------------CCA ....((.((((((((((..((....))..)))))))))).)).(((((.........)))))..(((.(..(((....)))..------))-------------)). ( -42.60) >consensus AAGCGGCAGCUGGCCAAAGUCGAAGGAAGUUGGUUGGCUGCCAUGUCAACGGCUAAAUGACACUGGCACUGGGAAUGGAAAUA______GGAA________GGAAUA ..(((((((((((((((..((....))..))))))))))))).(((((.........)))))...))........................................ (-33.49 = -33.38 + -0.11)

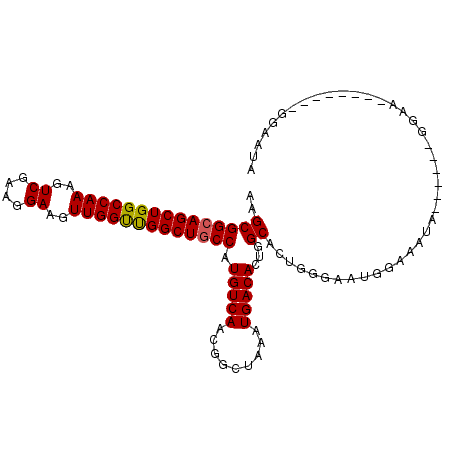

| Location | 23,921,592 – 23,921,693 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 85.98 |
| Mean single sequence MFE | -24.45 |
| Consensus MFE | -22.10 |
| Energy contribution | -21.97 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.63 |
| Structure conservation index | 0.90 |
| SVM decision value | 2.06 |
| SVM RNA-class probability | 0.986880 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 23921592 101 - 27905053 UAUUCCUAUUCCUAUUCC------UAUUUCCAUUCCCAGUGCCAGUGUCAUUUAGCCGUUGACAUGGCAGCCAACCAACUUCCUUCGACUUUGGCCAGCUGCCGCUU ..................------................((..((((((.........))))))((((((...((((..((....))..))))...)))))))).. ( -22.30) >DroSec_CAF1 123236 89 - 1 UAU------------UCC------UAUUUCCAUUCCCAGUGCCAGUGUCAUUUAGCCGUUGACAUGGCAGCCAACCAACUUCCUUCGACUUUGGCCAGCUGCCGCUU ...------------...------................((..((((((.........))))))((((((...((((..((....))..))))...)))))))).. ( -22.30) >DroSim_CAF1 124970 95 - 1 UAUUCC------UAUUCC------UAUUUCCAUUCCCAGUGCCAGUGUCAUUUAGCCGUUGACAUGGCAGCCAACCAACUUCCUUCGACUUUGGCCAGCUGCCGCUU ......------......------................((..((((((.........))))))((((((...((((..((....))..))))...)))))))).. ( -22.30) >DroEre_CAF1 126663 107 - 1 UAUUCCUAUUCCUAUUUCCAUUCCCAUUCCCAUUCCCAGUGCCAGUGUCAUUUAGCCGUUGACAUGGCAGCCAACCAACUUCCUUCGACUUUGGCCAGCUGCCGCUU ........................................((..((((((.........))))))((((((...((((..((....))..))))...)))))))).. ( -22.30) >DroYak_CAF1 130461 95 - 1 UGUUCCUA------UUUC------CAUUCCCAUUCCCAGUGCCAGUGUCAUUUAGCCGUUGACAUGGCAGCCAACCAACUUCCUUCGACUUUGGCCAGCUGCCGCUU ........------....------................((..((((((.........))))))((((((...((((..((....))..))))...)))))))).. ( -22.30) >DroAna_CAF1 130966 88 - 1 UGG-------------CC------CAGGACCCAUCCCAGUGCCAGUGUCAUUUAGCCGUUGACAUGGUAGCCGGCCAACUUCCUUCGACUUUGGCCAGCUGCCGCUU (((-------------((------..(((....)))..).))))((((((.........))))))((((((.((((((..((....))..)))))).)))))).... ( -35.20) >consensus UAUUCC________UUCC______CAUUUCCAUUCCCAGUGCCAGUGUCAUUUAGCCGUUGACAUGGCAGCCAACCAACUUCCUUCGACUUUGGCCAGCUGCCGCUU ........................................((..((((((.........))))))((((((...((((..((....))..))))...)))))))).. (-22.10 = -21.97 + -0.14)



| Location | 23,921,619 – 23,921,712 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 87.10 |
| Mean single sequence MFE | -12.42 |
| Consensus MFE | -12.20 |
| Energy contribution | -12.20 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.31 |
| Structure conservation index | 0.98 |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.679756 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 23921619 93 - 27905053 CCAAUACUAAGACCAUUCCUAUUCCUAUUCCUAUUCC------UAUUUCCAUUCCCAGUGCCAGUGUCAUUUAGCCGUUGACAUGGCAGCCAACCAACU .....................................------...............(((((.(((((.........))))))))))........... ( -12.20) >DroSec_CAF1 123263 81 - 1 CCAAUACUAAGACCAUUCCUAU------------UCC------UAUUUCCAUUCCCAGUGCCAGUGUCAUUUAGCCGUUGACAUGGCAGCCAACCAACU ......................------------...------...............(((((.(((((.........))))))))))........... ( -12.20) >DroSim_CAF1 124997 87 - 1 CCAAUACUAAGACCAUUCCUAUUCC------UAUUCC------UAUUUCCAUUCCCAGUGCCAGUGUCAUUUAGCCGUUGACAUGGCAGCCAACCAACU .........................------......------...............(((((.(((((.........))))))))))........... ( -12.20) >DroEre_CAF1 126690 99 - 1 CCAAUACUGAGAUCAUUCCUAUUCCUAUUCCUAUUUCCAUUCCCAUUCCCAUUCCCAGUGCCAGUGUCAUUUAGCCGUUGACAUGGCAGCCAACCAACU .....((((.((........................................)).))))((((.(((((.........)))))))))............ ( -13.30) >DroYak_CAF1 130488 87 - 1 CCAAUACUUAGACCAUUCCUGUUCCUA------UUUC------CAUUCCCAUUCCCAGUGCCAGUGUCAUUUAGCCGUUGACAUGGCAGCCAACCAACU ...........................------....------...............(((((.(((((.........))))))))))........... ( -12.20) >consensus CCAAUACUAAGACCAUUCCUAUUCCUA____UAUUCC______UAUUUCCAUUCCCAGUGCCAGUGUCAUUUAGCCGUUGACAUGGCAGCCAACCAACU ..........................................................(((((.(((((.........))))))))))........... (-12.20 = -12.20 + 0.00)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:22:40 2006