
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 23,908,712 – 23,908,830 |
| Length | 118 |
| Max. P | 0.803887 |

| Location | 23,908,712 – 23,908,830 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.43 |
| Mean single sequence MFE | -38.41 |
| Consensus MFE | -23.97 |
| Energy contribution | -23.87 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.28 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.619477 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
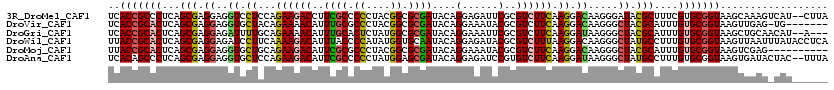
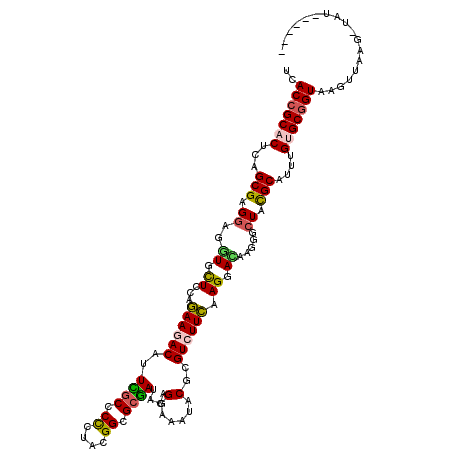
>3R_DroMel_CAF1 23908712 118 + 27905053 UCACCGCCCUCAGCGAGGAGGUCCUCCAGAAGACCUUCGCCCCCUACGGCGCGAUACAGGAGAUUCGCGUCUUCAAGGACAAGGGAUACGCUUUCGUGCGGUAAGCAAAGUCAU--CUUA ..((((((...((((.(((((((........))))))).((((((..(((((((..........)))))))....)))....)))...))))...).)))))............--.... ( -38.80) >DroVir_CAF1 105013 112 + 1 UCACCGCACUCAGCGAGGAGGUGCUACAGAAAACAUUUGCGCCCUACGGCGCGAUACAGGAAAUACGCGUCUUCAAGGACAAGGGCUACGCAUUUGUGCGGUAAGUUGAG-UG------- ......(((((((((((((.(((.............(((((((....)))))))...........))).)))))..(.(((((.((...)).))))).).....))))))-))------- ( -37.25) >DroGri_CAF1 75329 115 + 1 UCACCGCACUCAGCGAGGAGAUUUUGCAGAAAACAUUUGCACUCUAUGGCGCGAUACAGGAAAUUCGCGUCUUCAAGGAUAAGGGCUACGCAUUUGUGCGGUAAGCUGCAACAU--A--- ..(((((((...(((.((((....((((((.....))))))))))..(((((((..........))))))).................)))....)))))))............--.--- ( -34.50) >DroWil_CAF1 145530 120 + 1 UUACCGCACUCAGCGAGGAGAUCCUUCAAAAGACAUUUACCCCAUAUGGUGCAAUACAGGAGAUACGCGUCUUUAAGGACAAGGGCUAUGCCUUUGUGCGGUAAGUUAAUUUAUACCUCA (((((((((((......))).(((((..((((((....(((......)))((..............)))))))))))))(((((((...)))))))))))))))................ ( -33.14) >DroMoj_CAF1 117319 110 + 1 UUACCGCACUCAGCGAGGAGGUGCUGCAGAAGACAUUCGCGCCCUACGGCGCGAUACAGGAAAUACGCGUCUUCAAGGACAAGGGCUACGCAUUUGUGCGGUAAGUCGAG---------- (((((((((...(((.((..((.((...((((((..(((((((....)))))))....(......)..)))))).)).)).....)).)))....)))))))))......---------- ( -43.60) >DroAna_CAF1 117510 118 + 1 UCACAGCCCUCAGCGAGGAGGUGCUCCAGAAGACAUUCGCCCCCUAUGGAGCGAUACAGGAGAUCCGUGUCUUCAAGGAUAAGGGCUAUGCCUUUGUGCGGUAAGUGAUACUAC--UUUA (((((((((((......)))).)))((.((((((((((((.((....)).))))....((....))))))))))..(.((((((((...)))))))).)))...))))......--.... ( -43.20) >consensus UCACCGCACUCAGCGAGGAGGUGCUCCAGAAGACAUUCGCCCCCUACGGCGCGAUACAGGAAAUACGCGUCUUCAAGGACAAGGGCUACGCAUUUGUGCGGUAAGUUAAG_UAU______ ..(((((((...(((.((..((.((...((((((..((((.((....)).))))....(......)..)))))).)).)).....)).)))....))))))).................. (-23.97 = -23.87 + -0.11)
| Location | 23,908,712 – 23,908,830 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.43 |
| Mean single sequence MFE | -38.41 |
| Consensus MFE | -27.72 |
| Energy contribution | -28.00 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.803887 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
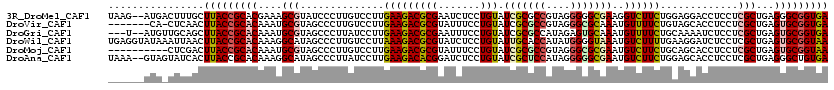
>3R_DroMel_CAF1 23908712 118 - 27905053 UAAG--AUGACUUUGCUUACCGCACGAAAGCGUAUCCCUUGUCCUUGAAGACGCGAAUCUCCUGUAUCGCGCCGUAGGGGGCGAAGGUCUUCUGGAGGACCUCCUCGCUGAGGGCGGUGA ....--..........((((((((((....)))...((((..(((((..(.(((((..(....)..))))))..)))))((((((((((((....)))))))..)))))))))))))))) ( -42.80) >DroVir_CAF1 105013 112 - 1 -------CA-CUCAACUUACCGCACAAAUGCGUAGCCCUUGUCCUUGAAGACGCGUAUUUCCUGUAUCGCGCCGUAGGGCGCAAAUGUUUUCUGUAGCACCUCCUCGCUGAGUGCGGUGA -------..-......(((((((((....(((.((....(((....(((((((.((((.....)))).(((((....)))))...)))))))....)))....)))))...))))))))) ( -35.50) >DroGri_CAF1 75329 115 - 1 ---U--AUGUUGCAGCUUACCGCACAAAUGCGUAGCCCUUAUCCUUGAAGACGCGAAUUUCCUGUAUCGCGCCAUAGAGUGCAAAUGUUUUCUGCAAAAUCUCCUCGCUGAGUGCGGUGA ---.--..........(((((((((....(((.((...........((((((((((..........))))(((.....).))....))))))...........)))))...))))))))) ( -28.65) >DroWil_CAF1 145530 120 - 1 UGAGGUAUAAAUUAACUUACCGCACAAAGGCAUAGCCCUUGUCCUUAAAGACGCGUAUCUCCUGUAUUGCACCAUAUGGGGUAAAUGUCUUUUGAAGGAUCUCCUCGCUGAGUGCGGUAA .................((((((((.((((......))))(((((((((((((..((((((.(((........))).))))))..)))))))..))))))...........)))))))). ( -35.30) >DroMoj_CAF1 117319 110 - 1 ----------CUCGACUUACCGCACAAAUGCGUAGCCCUUGUCCUUGAAGACGCGUAUUUCCUGUAUCGCGCCGUAGGGCGCGAAUGUCUUCUGCAGCACCUCCUCGCUGAGUGCGGUAA ----------.......((((((((....(((.((....(((....(((((((((.......))).(((((((....)))))))..))))))....)))....)))))...)))))))). ( -41.00) >DroAna_CAF1 117510 118 - 1 UAAA--GUAGUAUCACUUACCGCACAAAGGCAUAGCCCUUAUCCUUGAAGACACGGAUCUCCUGUAUCGCUCCAUAGGGGGCGAAUGUCUUCUGGAGCACCUCCUCGCUGAGGGCUGUGA ....--(..(((.....)))..).......((((((((((((((..((((((((((.....)))).(((((((....)))))))..)))))).)))((........))))))))))))). ( -47.20) >consensus ______AUA_CUCAACUUACCGCACAAAUGCGUAGCCCUUGUCCUUGAAGACGCGUAUCUCCUGUAUCGCGCCAUAGGGGGCAAAUGUCUUCUGCAGCACCUCCUCGCUGAGUGCGGUGA ................(((((((((....(((..............(((((((((.......))).(((((((....)))))))..)))))).............)))...))))))))) (-27.72 = -28.00 + 0.28)
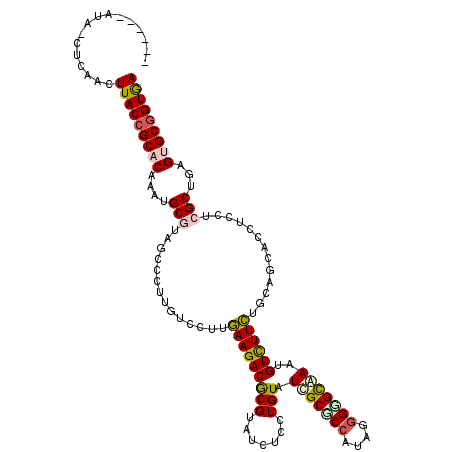

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:22:35 2006