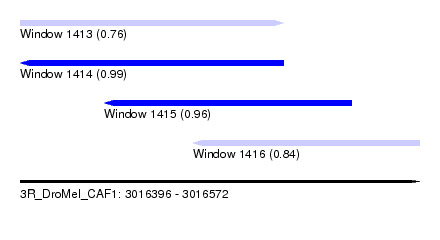
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 3,016,396 – 3,016,572 |
| Length | 176 |
| Max. P | 0.986064 |

| Location | 3,016,396 – 3,016,512 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 77.18 |
| Mean single sequence MFE | -16.09 |
| Consensus MFE | -12.04 |
| Energy contribution | -12.85 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.13 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.764448 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3016396 116 + 27905053 AAUAAUAUUUAACAGC-AUUCCUAUUGUUUUUCCUAAAUCUUUUCAGCGAUUUAUGCAAUAUCUUGCCUCUAGCAAUUCUACAUUCAAUCUCAGCUCCUUUCUGGACAAAGGAACUG ..............((-(....((((((......((((((........)))))).))))))...)))........................(((.((((((......)))))).))) ( -15.50) >DroVir_CAF1 5089 105 + 1 AAUCAGUAUAAA---CUUGUU-UUG--------CUCUCUUUUUGCAGCGUUUUAUGCAAUAUCUCGCAUCAAGCAACUCAACAUUCAAUCUCAGCUCCUUUCUGGACAAAGGAACUG ............---((((..-.((--------(.......(((((........)))))......))).))))..................(((.((((((......)))))).))) ( -17.72) >DroPse_CAF1 3684 106 + 1 AAUCAUCAUU-----C-UCUCUCAA--UUUU---UGUAUCUUUGCAGCGCUUCAUGCAAUAUUUAGCAUCAAGCAAUUCUACAUUCAAUCUCAGCUCCUUUCUGGACAAAGGAACUG ..........-----.-........--...(---((((....))))).((((.((((........)))).)))).................(((.((((((......)))))).))) ( -18.60) >DroGri_CAF1 4125 102 + 1 AAUCAAUUCUA------U-UCUUUU--------CUUUCUUUAUGCAGCGUUUCAUGCAAUAUCUCGCAUCGAGCAAUUCUACAUUCAAUCUCAGCUCCUUUCUGGACAAAGGAACUG ...........------.-......--------...............((((.((((........)))).)))).................(((.((((((......)))))).))) ( -14.70) >DroYak_CAF1 2924 93 + 1 AAUAAUAUUU------------------------AAAAUCUUUCCAGCGGUUUAUGCAAUAUCUUGCCUCUAGCAAUUCGACAUUCAAUCUCAGCUCCUUUCUGGAUAAAGGAACUG ..........------------------------............(((.....)))......((((.....))))...............(((.((((((......)))))).))) ( -11.40) >DroPer_CAF1 3732 106 + 1 AAUCAUCAUU-----C-UCUCUCAA--UUUU---UGUAUCUUUGCAGCGCUUCAUGCAAUAUUUAGCAUCAAGCAAUUCUACAUUCAAUCUCAGCUCCUUUCUGGACAAAGGAACUG ..........-----.-........--...(---((((....))))).((((.((((........)))).)))).................(((.((((((......)))))).))) ( -18.60) >consensus AAUCAUAAUU_____C_U_UCUUAA_________UAUAUCUUUGCAGCGCUUCAUGCAAUAUCUAGCAUCAAGCAAUUCUACAUUCAAUCUCAGCUCCUUUCUGGACAAAGGAACUG ................................................((((.((((........)))).)))).................(((.((((((......)))))).))) (-12.04 = -12.85 + 0.81)

| Location | 3,016,396 – 3,016,512 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 77.18 |
| Mean single sequence MFE | -26.60 |
| Consensus MFE | -17.83 |
| Energy contribution | -18.38 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.62 |
| Structure conservation index | 0.67 |
| SVM decision value | 2.03 |
| SVM RNA-class probability | 0.986064 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3016396 116 - 27905053 CAGUUCCUUUGUCCAGAAAGGAGCUGAGAUUGAAUGUAGAAUUGCUAGAGGCAAGAUAUUGCAUAAAUCGCUGAAAAGAUUUAGGAAAAACAAUAGGAAU-GCUGUUAAAUAUUAUU ((((((((((......)))))))))).((((..((((((..((((.....))))....)))))).)))).(((((....))))).......(((((....-.))))).......... ( -26.90) >DroVir_CAF1 5089 105 - 1 CAGUUCCUUUGUCCAGAAAGGAGCUGAGAUUGAAUGUUGAGUUGCUUGAUGCGAGAUAUUGCAUAAAACGCUGCAAAAAGAGAG--------CAA-AACAAG---UUUAUACUGAUU ((((((((((......))))))))))((..(((((.(((..((((((.((((((....))))))......((......)).)))--------)))-..))))---))))..)).... ( -28.00) >DroPse_CAF1 3684 106 - 1 CAGUUCCUUUGUCCAGAAAGGAGCUGAGAUUGAAUGUAGAAUUGCUUGAUGCUAAAUAUUGCAUGAAGCGCUGCAAAGAUACA---AAAA--UUGAGAGA-G-----AAUGAUGAUU ((((((((((......))))))))))........(((((....((((.((((........)))).)))).)))))........---....--........-.-----.......... ( -29.20) >DroGri_CAF1 4125 102 - 1 CAGUUCCUUUGUCCAGAAAGGAGCUGAGAUUGAAUGUAGAAUUGCUCGAUGCGAGAUAUUGCAUGAAACGCUGCAUAAAGAAAG--------AAAAGA-A------UAGAAUUGAUU ((((((((((......)))))))))).......((((((......((.((((((....))))))))....))))))........--------......-.------........... ( -25.00) >DroYak_CAF1 2924 93 - 1 CAGUUCCUUUAUCCAGAAAGGAGCUGAGAUUGAAUGUCGAAUUGCUAGAGGCAAGAUAUUGCAUAAACCGCUGGAAAGAUUUU------------------------AAAUAUUAUU ((((((((((......))))))))))......(((((.(((((.((((..((((....))))........))))...))))).------------------------..)))))... ( -21.30) >DroPer_CAF1 3732 106 - 1 CAGUUCCUUUGUCCAGAAAGGAGCUGAGAUUGAAUGUAGAAUUGCUUGAUGCUAAAUAUUGCAUGAAGCGCUGCAAAGAUACA---AAAA--UUGAGAGA-G-----AAUGAUGAUU ((((((((((......))))))))))........(((((....((((.((((........)))).)))).)))))........---....--........-.-----.......... ( -29.20) >consensus CAGUUCCUUUGUCCAGAAAGGAGCUGAGAUUGAAUGUAGAAUUGCUUGAUGCAAGAUAUUGCAUAAAACGCUGCAAAGAUACA_________AUAAGA_A_G_____AAAAAUGAUU ((((((((((......))))))))))((.....((((((..((((.....))))....))))))......))............................................. (-17.83 = -18.38 + 0.56)
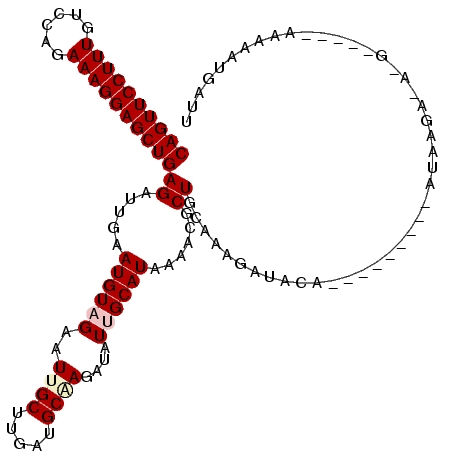
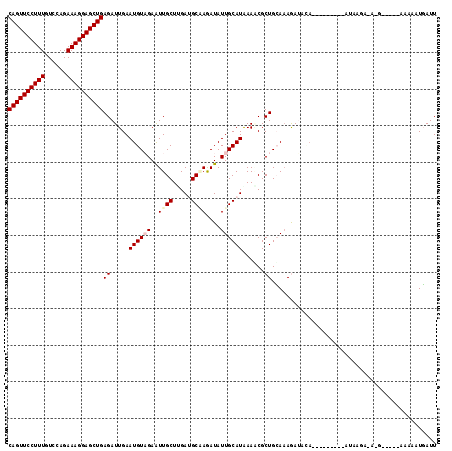
| Location | 3,016,433 – 3,016,542 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 77.80 |
| Mean single sequence MFE | -28.79 |
| Consensus MFE | -18.95 |
| Energy contribution | -18.95 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.66 |
| SVM decision value | 1.46 |
| SVM RNA-class probability | 0.955501 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3016433 109 - 27905053 --------CUUA--AAAGGGAGGCUUCGUUAUACCUUGUACAGUUCCUUUGUCCAGAAAGGAGCUGAGAUUGAAUGUAGAAUUGCUAGAGGCAAGAUAUUGCAUAAAUCGCUGAAAAGA --------....--.((((..............))))...((((((((((......)))))))))).((((..((((((..((((.....))))....)))))).)))).((....)). ( -27.24) >DroVir_CAF1 5115 117 - 1 GUUUUUUGUUUGUGAAAUUCAUUCG--GUUAUACCUUGCACAGUUCCUUUGUCCAGAAAGGAGCUGAGAUUGAAUGUUGAGUUGCUUGAUGCGAGAUAUUGCAUAAAACGCUGCAAAAA ..(((((((..(((..(((((.(((--(.....)).....((((((((((......)))))))))).)).))))).............((((((....))))))....))).))))))) ( -32.60) >DroPse_CAF1 3711 110 - 1 --------UUAGU-UAAGGGCAAUAUCUUUAUACCUUGCACAGUUCCUUUGUCCAGAAAGGAGCUGAGAUUGAAUGUAGAAUUGCUUGAUGCUAAAUAUUGCAUGAAGCGCUGCAAAGA --------(((((-(((((...(((....))).))))...((((((((((......)))))))))).)))))).(((((....((((.((((........)))).)))).))))).... ( -33.80) >DroGri_CAF1 4148 112 - 1 -U------UUAGUGAAAUGCAUUCGGGGUUAUACCUUGCACAGUUCCUUUGUCCAGAAAGGAGCUGAGAUUGAAUGUAGAAUUGCUCGAUGCGAGAUAUUGCAUGAAACGCUGCAUAAA -.------.(((((...(((((((((((.....))))...((((((((((......)))))))))).....))))))).......((.((((((....))))))))..)))))...... ( -34.30) >DroWil_CAF1 5000 99 - 1 --------------------CAAUAGUUUUAUACCUUGCACAGUUCCUUUAUCCAAAAAGGAGCUGAGAUUGAAUGUUGAAUUGCUCGAUGCCAGAUAUUGCAUAAAUCUCUGUAAAAG --------------------...............(((((.(((((((((......)))))))))((((((..((((...(((((.....))..)))...)))).)))))))))))... ( -22.30) >DroYak_CAF1 2938 103 - 1 --------UUUA--AAAAAUA------UUUAUACCUUGUACAGUUCCUUUAUCCAGAAAGGAGCUGAGAUUGAAUGUCGAAUUGCUAGAGGCAAGAUAUUGCAUAAACCGCUGGAAAGA --------....--.......------......((..((.((((((((((......))))))))))....((((((((...((((.....)))))))))).))......)).))..... ( -22.50) >consensus ________UUUG__AAAGGCAAACA__GUUAUACCUUGCACAGUUCCUUUGUCCAGAAAGGAGCUGAGAUUGAAUGUAGAAUUGCUAGAUGCAAGAUAUUGCAUAAAACGCUGCAAAAA ....................................(((.((((((((((......))))))))))......(((((....((((.....)))).))))))))................ (-18.95 = -18.95 + 0.00)
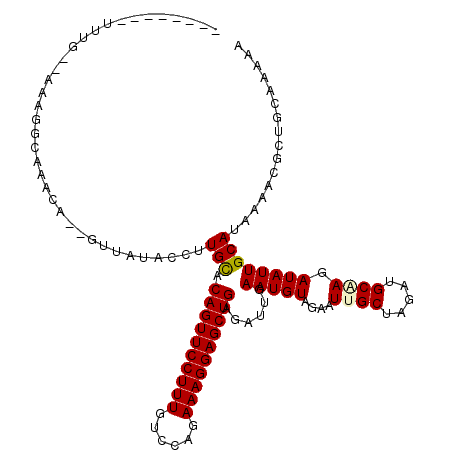
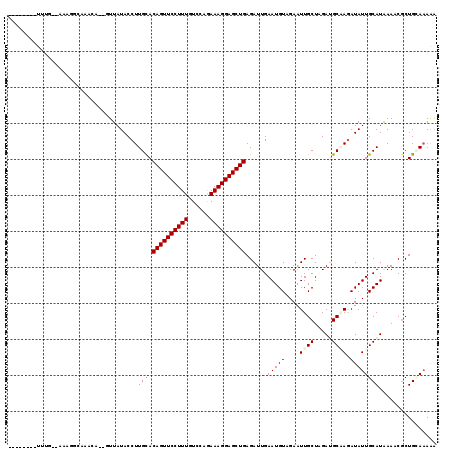
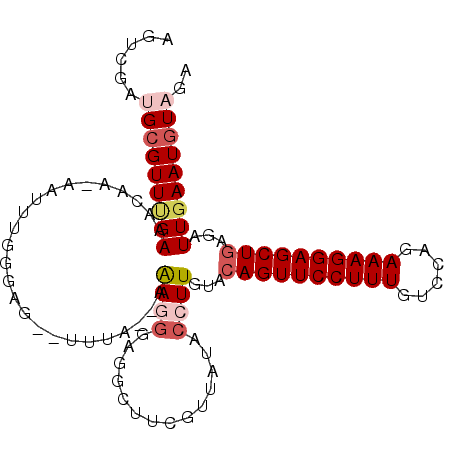
| Location | 3,016,472 – 3,016,572 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 79.79 |
| Mean single sequence MFE | -26.24 |
| Consensus MFE | -17.14 |
| Energy contribution | -18.14 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.75 |
| SVM RNA-class probability | 0.840009 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3016472 100 - 27905053 AGUCGAUGGGUUUGAAACAA--AUUUGGGAGG-CUUA--AAAGGGAGGCUUCGUUAUACCUUGUACAGUUCCUUUGUCCAGAAAGGAGCUGAGAUUGAAUGUAGA ..(((((((((.(((..((.--...)).((((-(((.--......))))))).))).))))....((((((((((......))))))))))..)))))....... ( -26.80) >DroGri_CAF1 4187 94 - 1 UGUUGUCGCGUU-GAUCGAG-AU---------UUUAGUGAAAUGCAUUCGGGGUUAUACCUUGCACAGUUCCUUUGUCCAGAAAGGAGCUGAGAUUGAAUGUAGA .....((((...-(((....-))---------)...))))..(((((((((((.....))))...((((((((((......)))))))))).....))))))).. ( -28.10) >DroSec_CAF1 3310 101 - 1 AGUCAAUGCGUUUGAAACAAAAAUUUGGGAG--UUUA--AGAGGGUGGCUUCGUUAUACCUUGUACAGUUCCUUUGUCCAGAAAGGAGCUAAGAUUGAAUGUAGA ......((((((..(...............(--(...--.(((((((((...))))).))))..))(((((((((......)))))))))....)..)))))).. ( -22.90) >DroSim_CAF1 3368 101 - 1 AGUCAAUGCGUUUGAAACAAGAAUUUGGGAG--UUUA--AGAGGGUGGCUUCGUUAUACCUUGUACAGUUCCUUUGUCCAGAAAGGAGCUGAGAUUGAAUGUAGA ......((((((..(.(((((......((((--((..--.......)))))).......))))).((((((((((......))))))))))...)..)))))).. ( -27.32) >DroEre_CAF1 3435 100 - 1 CGUCGAUGCGUUUGAAGCAA-AUUUUGGACA--UUUA--AAAGGGACGCUUCGUUAUACCUUGUACAGUUCCUUUAUCCAGAAAGGAGCUGAGAUUGAAUGUAGA ......(((.......))).-.......(((--((((--(((((((((...))))...))))...((((((((((......))))))))))...))))))))... ( -25.50) >DroYak_CAF1 2977 94 - 1 CGUCGAUGCGUUCGAAACAA-AUUUUGGAUA--UUUA--AAAAAUA------UUUAUACCUUGUACAGUUCCUUUAUCCAGAAAGGAGCUGAGAUUGAAUGUCGA ..(((..((((((((.((((-....((((((--((..--...))))------))))....)))).((((((((((......))))))))))...))))))))))) ( -26.80) >consensus AGUCGAUGCGUUUGAAACAA_AAUUUGGGAG__UUUA__AAAGGGAGGCUUCGUUAUACCUUGUACAGUUCCUUUGUCCAGAAAGGAGCUGAGAUUGAAUGUAGA ......(((((((((.........................((((..............))))...((((((((((......))))))))))...))))))))).. (-17.14 = -18.14 + 1.00)

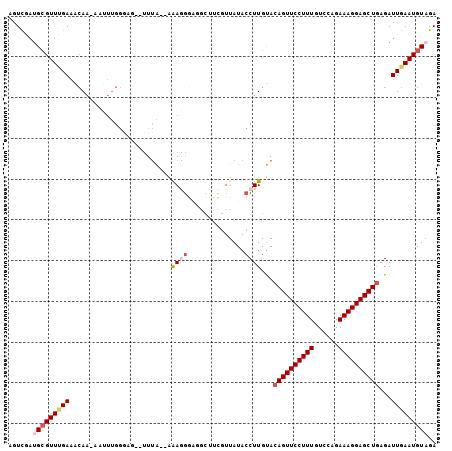
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:54:45 2006