
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 23,869,237 – 23,869,356 |
| Length | 119 |
| Max. P | 0.687113 |

| Location | 23,869,237 – 23,869,356 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 79.32 |
| Mean single sequence MFE | -27.67 |
| Consensus MFE | -18.34 |
| Energy contribution | -18.78 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.687113 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 23869237 119 - 27905053 CCAUUUCCAUGGCACGCAGCGCGGGCAGGUUCCUGGGCAAUUUGCCAUUCUGCAAUGGAGAGCGAACGAUGCGAUGGACCAUUAAAAUUAGACAAUAAAUAGAUAAAGAUGAAAAUAAA .....(((((.(((((...(((.(((((((((....).)))))))).((((.....)))).)))..)).))).)))))......................................... ( -28.90) >DroVir_CAF1 66049 95 - 1 CCAUUUCCAUGGCACGCAAUGCGGGCAGAUUGCGGGGCAAUUUGCCGUUCUGCAAUGGA------GACAAGCGUUGGCAUAUUAAAAUUAGACA-----UGGA------------UA-A .....((((((((..(((..((((.((((((((...))))))))))))..)))..((..------..)).))(....)..............))-----))))------------..-. ( -31.30) >DroPse_CAF1 81419 112 - 1 CCAUUUCCAUGGCACGCAGGGCUGGCAGGUUCCUGGGCAAUUUGCCAUUCUGCAAUGGA------GCCAAGUGAUGGACCAUUAAAAUUACACA-UAAAUAGAUAAGGAUAAAAAUAAG .....(((.......((((((.((((((((((....).)))))))))))))))(((((.------.(((.....))).)))))...........-...........))).......... ( -29.10) >DroWil_CAF1 105125 110 - 1 CCAUUUCCAUGGCACGCAAUGCGGGCAGGUUUCUGGGCAAUUUGCCAUUCUGCAAUGAA------GUUAAGUGAUGGCAUAUGAAAAUUAAACA-UAAAUAGAUAAAAC-AAAACGAA- .(((..((((.((.(((...))).(((((......(((.....)))..)))))......------.....)).))))...)))...........-..............-........- ( -21.40) >DroAna_CAF1 82312 115 - 1 CCAUUUCCAUUGCACGCAAUGCCGGCAGGCUCCUGGGCAAUUUGCCAUUCUGCAACGAA-AG--AGUCCAGCGAUGGACCAUCAACAUUAGACA-UAAAUAUAUAAAGACAAAAAUAAA .....((((((((.......(((....)))...(((((...((((......))))(...-.)--.)))))))))))))................-........................ ( -27.30) >DroPer_CAF1 81212 112 - 1 CCAUUUCCAUGGCACGCAGGGCGGGCAGGUUCCUGGGCAAUUUGCCAUUCUGCAAUGGA------GCCAAGUGAUGGACCAUUAAAAUUACACA-UAAAUAGAUAAGGAUAAAAAUAAG .....(((.((((.(.((..((((((((....)))(((.....)))..)))))..))).------)))).(((((...........)))))...-...........))).......... ( -28.00) >consensus CCAUUUCCAUGGCACGCAAUGCGGGCAGGUUCCUGGGCAAUUUGCCAUUCUGCAAUGGA______GCCAAGCGAUGGACCAUUAAAAUUAGACA_UAAAUAGAUAAAGAUAAAAAUAAA .....(((((.((.(((...))).(((((......(((.....)))..))))).................)).)))))......................................... (-18.34 = -18.78 + 0.45)

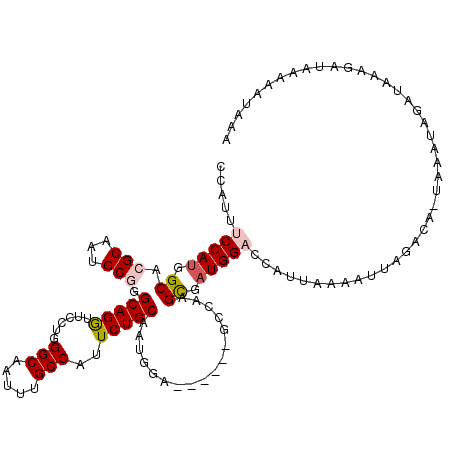
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:22:20 2006