
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 3,006,341 – 3,006,568 |
| Length | 227 |
| Max. P | 0.995275 |

| Location | 3,006,341 – 3,006,454 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 74.71 |
| Mean single sequence MFE | -29.07 |
| Consensus MFE | -19.82 |
| Energy contribution | -20.86 |
| Covariance contribution | 1.04 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.68 |
| SVM decision value | 1.47 |
| SVM RNA-class probability | 0.956535 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
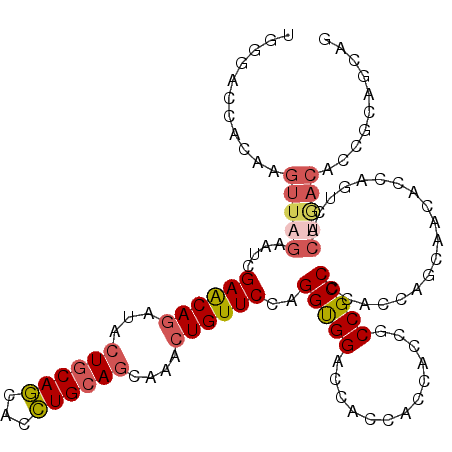
>3R_DroMel_CAF1 3006341 113 + 27905053 UGGGACCACAAGUUAGAAUCGAACAGAUACUGCAGCACCUGCAGCAAACUGUUCCAGGUGGACCACCACCACCUCCGCCCCACCAGCAACACCAAUCCCUGACACCGCAGCUG ((((................((((((...((((((...))))))....))))))..(((((....)))))........)))).((((......................)))) ( -30.05) >DroSec_CAF1 52695 104 + 1 UGGGACCGCAAGUUAGAAUCGAACAGAUACUGCAGCACCUGCAGCAAACUGUUCCAGGUGGACCACCACCACCGCCGCCCCACCAGCAACACCAGUCACUGACA--------- ((((.(.((...........((((((...((((((...))))))....))))))..(((((....)))))...)).).)))).(((..((....))..)))...--------- ( -33.50) >DroSim_CAF1 53123 104 + 1 UGGGACCACAAGUUAGAAUCGAACAGAUACUGCAGCACCUGCAGCAAACUGUUCCAGGUGGAC---------CGCCGCCCAACCAGCAACACCAGUCACUGACACCGCAGCAG ..((.((((..(.......)((((((...((((((...))))))....))))))...)))).)---------)((.((.....(((..((....))..))).....)).)).. ( -27.40) >DroWil_CAF1 56504 92 + 1 UGCAGCCGCAUGUCAGGAUUGAGCAGAUUCUACAACAUUUGCAACAAAGUGUUCCAGGCGGACAGCCACCACCUCCGCCG--------ACA-------------UCACAACAG .((....))(((((((((((.....))))))..(((((((......)))))))...((((((...........)))))))--------)))-------------)........ ( -23.00) >DroYak_CAF1 61012 110 + 1 UGGGACCACAAGUUCGAAUCGAACAGAUACUGCAGCACCUGCAGCAAACUGUUCCAGGUGGACCACC---ACCGCCGCCCCAUCAGCAGCACCAGUCUCUAACACCGCAGCAG .(((((..............((((((...((((((...))))))....))))))..(((((....))---)))((.((.......)).))....))))).............. ( -31.40) >consensus UGGGACCACAAGUUAGAAUCGAACAGAUACUGCAGCACCUGCAGCAAACUGUUCCAGGUGGACCACCACCACCGCCGCCCCACCAGCAACACCAGUCACUGACACCGCAGCAG ...........(((((....((((((...((((((...))))))....))))))..(((((.............)))))...................))))).......... (-19.82 = -20.86 + 1.04)

| Location | 3,006,341 – 3,006,454 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 74.71 |
| Mean single sequence MFE | -43.22 |
| Consensus MFE | -28.78 |
| Energy contribution | -30.18 |
| Covariance contribution | 1.40 |
| Combinations/Pair | 1.34 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.67 |
| SVM decision value | 2.56 |
| SVM RNA-class probability | 0.995275 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3006341 113 - 27905053 CAGCUGCGGUGUCAGGGAUUGGUGUUGCUGGUGGGGCGGAGGUGGUGGUGGUCCACCUGGAACAGUUUGCUGCAGGUGCUGCAGUAUCUGUUCGAUUCUAACUUGUGGUCCCA ..(((((((..((((...))))..)))).)))(((((.((((((((((....)))))..((((((..((((((((...)))))))).)))))).......)))).).))))). ( -46.70) >DroSec_CAF1 52695 104 - 1 ---------UGUCAGUGACUGGUGUUGCUGGUGGGGCGGCGGUGGUGGUGGUCCACCUGGAACAGUUUGCUGCAGGUGCUGCAGUAUCUGUUCGAUUCUAACUUGCGGUCCCA ---------..((((..((....))..))))((((((.((((((((((....)))))..((((((..((((((((...)))))))).)))))).......)).))).)))))) ( -50.50) >DroSim_CAF1 53123 104 - 1 CUGCUGCGGUGUCAGUGACUGGUGUUGCUGGUUGGGCGGCG---------GUCCACCUGGAACAGUUUGCUGCAGGUGCUGCAGUAUCUGUUCGAUUCUAACUUGUGGUCCCA ..(((((....((((..((....))..))))....)))))(---------(.((((.(.((((((..((((((((...)))))))).)))))).).........)))).)).. ( -42.00) >DroWil_CAF1 56504 92 - 1 CUGUUGUGA-------------UGU--------CGGCGGAGGUGGUGGCUGUCCGCCUGGAACACUUUGUUGCAAAUGUUGUAGAAUCUGCUCAAUCCUGACAUGCGGCUGCA ..(((((.(-------------(((--------((((((((((....))).)))))).(((.....(((..(((.((........)).))).)))))).)))))))))).... ( -27.00) >DroYak_CAF1 61012 110 - 1 CUGCUGCGGUGUUAGAGACUGGUGCUGCUGAUGGGGCGGCGGU---GGUGGUCCACCUGGAACAGUUUGCUGCAGGUGCUGCAGUAUCUGUUCGAUUCGAACUUGUGGUCCCA .....((((..((((...))))..))))...((((((.(((((---((....)))))..((((((..((((((((...)))))))).))))))...........)).)))))) ( -49.90) >consensus CUGCUGCGGUGUCAGUGACUGGUGUUGCUGGUGGGGCGGCGGUGGUGGUGGUCCACCUGGAACAGUUUGCUGCAGGUGCUGCAGUAUCUGUUCGAUUCUAACUUGUGGUCCCA ...........((((..((....))..)))).(((((.(((((((.......)))))..((((((..((((((((...)))))))).))))))...........)).))))). (-28.78 = -30.18 + 1.40)


| Location | 3,006,454 – 3,006,568 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 76.62 |
| Mean single sequence MFE | -35.45 |
| Consensus MFE | -28.18 |
| Energy contribution | -28.93 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.12 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.72 |
| SVM RNA-class probability | 0.833719 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3006454 114 + 27905053 CAUCCGC-AGCAGCAACAAAUUUCGCAGCAACAUCCUGGUCAUUAUGUGCCCAUCGUGCACAGUGGAGUGCCGCCACCGCCACCAGGACAUGGCAUUGCCAUUGUCGAUGGGCAA .......-.((.((..........)).))..................(((((((((...((((((((((((((...((.......))...))))))).)))))))))))))))). ( -38.90) >DroSec_CAF1 52799 114 + 1 CAUCCGC-AGCAGCAACCAACUCCGCAGCAACAUCCUGAUCAUUAUGUGCCCAUCGUGCACAGUGGAGUGCCGCCGCCGCCGCCAGGACAUGGCAUUGCCAUUGUCGAUGGGCAA .......-.((.((..........)).))..................(((((((((...((((((((((((((...((.......))...))))))).)))))))))))))))). ( -40.00) >DroSim_CAF1 53227 114 + 1 CAUCCGC-AGCAGCAACCAACUCCGCAGCAACAUCCUGGUCAUUAUGUGCCCAUCGUGCACAGUGGAGUGCCGCCGCCGCCGCCAGGACAUGGCAUUGCCGUUGUCGAUGGGCAA ((((.((-(((.((((...(((((((..((......)).......(((((.......))))))))))))...((((...((....))...)))).)))).))))).))))..... ( -39.30) >DroYak_CAF1 61122 108 + 1 CAGCAGCA-------ACAAGCUCCGCAGCAACAUCCGGGUCAUUAUGUGCCCAUCGUACACAGUGGAGUGCCACCGCCGCCACCAGGACAUGGCAUUGCCAUUGUCGAUGGGCAA ..(((((.-------....)))..)).....................(((((((((...((((((((((((((...((.......))...))))))).)))))))))))))))). ( -41.50) >DroMoj_CAF1 50951 101 + 1 ----AGC----AACAACAAAGACCU------CUGCCUCCUCAUUAUGUGCCCAUAGUGCACAGUGGGGUGCCACCACCGCCGCCGCCCCAUGGCAUUGCUAUAGUCGAUGGCCAG ----...----..............------.((((.........(((((.......)))))((((((((.(.........).))))))))))))..(((((.....)))))... ( -26.80) >DroAna_CAF1 45519 94 + 1 ---------------------ACAGCCCCAGCAUCCAGGACACUAUGUGCCCAUUGUGCACAGUGGAGUCCCGCCACCCCCACCAGCUCAUGGCAUUGCCAUUGUGGAUGGACAG ---------------------...((....)).((((((((.((((((((.......))))).))).))))........((((......(((((...))))).)))).))))... ( -26.20) >consensus CAUCCGC____AGCAACAAACUCCGCAGCAACAUCCUGGUCAUUAUGUGCCCAUCGUGCACAGUGGAGUGCCGCCACCGCCACCAGGACAUGGCAUUGCCAUUGUCGAUGGGCAA ...............................................(((((((((...((((((((((((((...((.......))...))))))).)))))))))))))))). (-28.18 = -28.93 + 0.75)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:54:41 2006