
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 23,868,352 – 23,868,451 |
| Length | 99 |
| Max. P | 0.661772 |

| Location | 23,868,352 – 23,868,451 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.74 |
| Mean single sequence MFE | -23.33 |
| Consensus MFE | -17.98 |
| Energy contribution | -18.48 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.08 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.661772 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 23868352 99 - 27905053 U-UUUCCGGC------------CGAGCCCCU--------GCCGGACAUUUAGUUUAUUUGAUGUUGCCAAAUGAAAAUUGUGACAUAAAAGGGACUAAGAACAUAAUUAACUAGUGUCCA .-..((((((------------.(.....).--------))))))..(((((((..(((.((((..(..(((....))))..)))).)))..)))))))..................... ( -24.40) >DroVir_CAF1 65038 96 - 1 GUGGUGC---------------CCCGCCCC---------UUAGGACAUUUAGUUUAUUUGAUGUUGCGAAAUGAAAAUUGUGACAUAAAAGGGACUAACAACAUAAUUAAGUAGUGUCCG ((((...---------------.))))...---------...((((((((((((..(((.((((..(((........)))..)))).)))..))))))..((........)).)))))). ( -24.90) >DroGri_CAF1 35007 98 - 1 GUCGUACAC-------------CCCGCCCA---------UGCGGACAUUUAGUUUAUUUGAUGUUGCGAAAUGAAAAUUGUGACAUAAAAGGGACUAACAACAUAAUUAAGUAGUGUCCA ...(.((((-------------.((((...---------.))))....((((((..(((.((((..(((........)))..)))).)))..))))))...............)))).). ( -23.80) >DroWil_CAF1 104081 93 - 1 C-CUUUUUGC------------UGAA--------------CCGCACAUUUAGUUUAUUUGAUGUUGCGAAAUGAAAAUUGUGACAUAAAAGGGACUAAGAACAUAAUUAAGUAGUGUCCA .-........------------....--------------..((((.(((((((..(((.((((..(((........)))..)))).)))..))))))).((........)).))))... ( -17.90) >DroMoj_CAF1 72291 111 - 1 GCUGUACAGCCACACCCCCACCGCCGCCCC---------UUAGGACAUUUAGUUUAUUUGAUGUUGCGAAAUGAAAAUUGUGACAUAAAAGGGACUAACAACAUAAUUAAGUAGUGUCCA (((....)))....................---------...((((((((((((..(((.((((..(((........)))..)))).)))..))))))..((........)).)))))). ( -23.40) >DroPer_CAF1 80291 107 - 1 A-GCCUCUGG------------CGAGCCCCUGUCGUCCGGCCGGACAUUUACUUUAUUUGAUGUGGCGAAAUGAAAAUUGUGACAUAAAAGGGACUAAGAACAUAAUUAAGUAGUGUCCG .-(((...((------------(((.......))))).)))((((((((.((((.(((..((((.((((........)))).))))..................))).)))))))))))) ( -25.55) >consensus G_CGUACAGC____________CCAGCCCC_________UCCGGACAUUUAGUUUAUUUGAUGUUGCGAAAUGAAAAUUGUGACAUAAAAGGGACUAACAACAUAAUUAAGUAGUGUCCA ..........................................((((((((((((..(((.((((..(((........)))..)))).)))..))))))...............)))))). (-17.98 = -18.48 + 0.50)

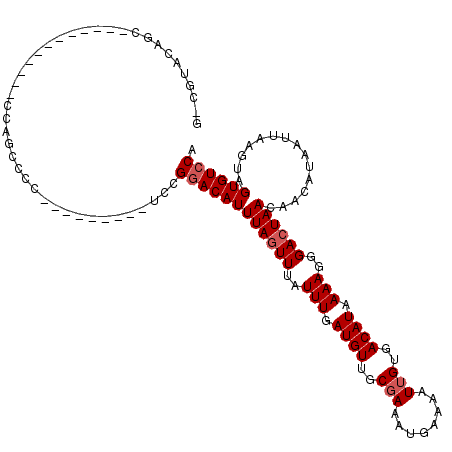
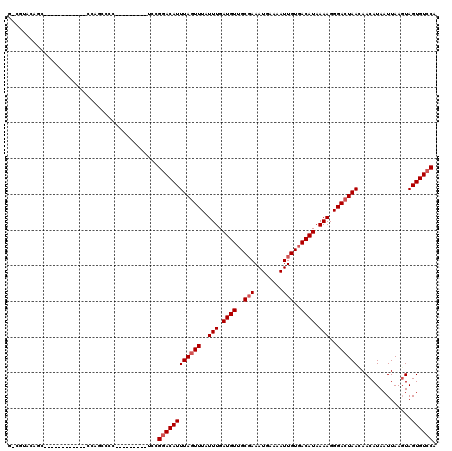
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:22:18 2006