| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 23,856,210 – 23,856,345 |
| Length | 135 |
| Max. P | 0.697482 |

| Location | 23,856,210 – 23,856,320 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 79.83 |
| Mean single sequence MFE | -35.85 |
| Consensus MFE | -23.53 |
| Energy contribution | -23.12 |
| Covariance contribution | -0.41 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.26 |
| Structure conservation index | 0.66 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.506510 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 23856210 110 - 27905053 GCAGGAUACCCACGAUUCCCAGUCCCGAGGCGGGACUGGUCGAUACUCCGGUCGACAGGUUCGCUAUUUUGGCCGCGACAGCAAGGUCAGCUUUAUUUAACUGCAAAUA-A ((((........(((..((((((((((...))))))))((((((......)))))).)).))).....((((((((....))..))))))..........)))).....-. ( -38.70) >DroVir_CAF1 51983 106 - 1 GCAGGAUGCCCAUUAAGCGCAGUCCCACGGCGGGACUGGCAGGCUGUCCAGUUGCUAGGUUGUCUAUUUUGGCGGCGUCUUC----UCUG-CUUGUUCAACUGCAAAAUAA ((((((((((......((.(((((((.....))))))))).((((....))))((((((........))))))))))))...----.)))-)((((......))))..... ( -36.60) >DroSec_CAF1 61757 110 - 1 GCAGGAUGCCCACGAUUCCCAGUCCCGAGGCGGGACUGGUCGAUACUCCGGUCGACAGGUUCGCUAUUUUGGCCGCGAGAAAAAGGUCAGCUAUAUUUAACUGAAAAUA-A ((.....))((.(((...(((((((((...))))))))))))...((((((((((..((....))...))))))).))).....))((((.((....)).)))).....-. ( -35.80) >DroSim_CAF1 61186 110 - 1 GCAGGAUGCCCACGAUUCCCAGUCCCGAGGCGGGACUGGUCGAUACUCCGGUCGACAGGUUCGCUAUCUUGGCCUCGACAGAAAGGUCAGCUAUAUUUAACUGAAAAUA-A ((((((((....(((..((((((((((...))))))))((((((......)))))).)).))).))))))(((((........))))).))..................-. ( -35.20) >DroEre_CAF1 65728 110 - 1 GCAGGAUGCCCACGAUACCCAGUCCCGCGGCGGGACUGGUCGAUACGCCGGUCGACAGGUUCGCUAUUUUGGCAGCGUCAGAUAGGUCAGCUAUAUUUAACUGAAAUUA-A .((((((((....((.(((((((((((...))))))))((((((......)))))).)))))(((.....))).)))))..((((.....))))......)))......-. ( -37.50) >DroYak_CAF1 63658 100 - 1 GCAGGAUGCCCACCAAUCCCAGUCCCGCGGCGGGACUGGUCGGUACUCCGGUCGACAGGUUCGCUAUUUUUGGCUCCUC----------UUUAUAUUUAGCUGAAAAUU-A ((.((....))(((....(((((((((...)))))))))((((....))))......)))..)).(((((..(((....----------.........)))..))))).-. ( -31.32) >consensus GCAGGAUGCCCACGAUUCCCAGUCCCGAGGCGGGACUGGUCGAUACUCCGGUCGACAGGUUCGCUAUUUUGGCCGCGACAGAAAGGUCAGCUAUAUUUAACUGAAAAUA_A .(((((((((.........(((((((.....)))))))((((((......)))))).))).....))))))........................................ (-23.53 = -23.12 + -0.41)

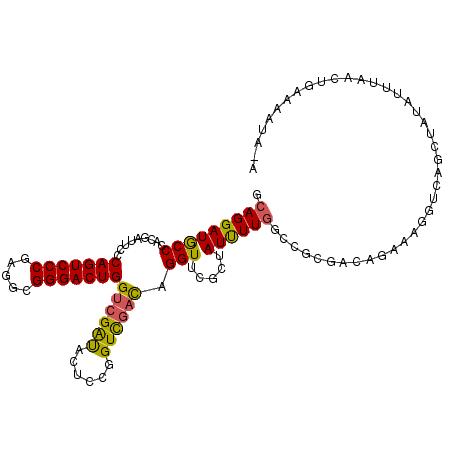

| Location | 23,856,241 – 23,856,345 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 86.22 |
| Mean single sequence MFE | -48.98 |
| Consensus MFE | -40.52 |
| Energy contribution | -41.97 |
| Covariance contribution | 1.45 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.697482 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 23856241 104 - 27905053 GGCGGCUGCCAAUGGAGCGAGCAGGGCAGGAUACCCACGAUUCCCAGUCCCGAGGCGGGACUGGUCGAUACUCCGGUCGACAGGUUCGCUAUUUUGGCCGCGAC ....((.(((((...(((((((.(((.......)))........((((((((...))))))))((((((......))))))..)))))))...))))).))... ( -48.00) >DroVir_CAF1 52010 104 - 1 GAUGGCUGCCACUGCAGUGCGCACGGCAGGAUGCCCAUUAAGCGCAGUCCCACGGCGGGACUGGCAGGCUGUCCAGUUGCUAGGUUGUCUAUUUUGGCGGCGUC (((((((((((......(((((..(((.....)))......)))))(((((.....)))))))))).))))))..(((((((((........)))))))))... ( -42.80) >DroSec_CAF1 61788 104 - 1 GGCGGCUGCCAAUGGAGCGAGCAGGGCAGGAUGCCCACGAUUCCCAGUCCCGAGGCGGGACUGGUCGAUACUCCGGUCGACAGGUUCGCUAUUUUGGCCGCGAG ....((.(((((...(((((((.((((.....))))........((((((((...))))))))((((((......))))))..)))))))...))))).))... ( -52.40) >DroSim_CAF1 61217 104 - 1 GGCGGCUGCCAAUGGAGCGAGCAGGGCAGGAUGCCCACGAUUCCCAGUCCCGAGGCGGGACUGGUCGAUACUCCGGUCGACAGGUUCGCUAUCUUGGCCUCGAC ..(((..(((((...(((((((.((((.....))))........((((((((...))))))))((((((......))))))..)))))))...))))).))).. ( -49.20) >DroEre_CAF1 65759 104 - 1 GGCGGCUGCCAAUGGAGUGAGCAGGGCAGGAUGCCCACGAUACCCAGUCCCGCGGCGGGACUGGUCGAUACGCCGGUCGACAGGUUCGCUAUUUUGGCAGCGUC ((((.(((((((...(((((((.((((.....))))........((((((((...))))))))((((((......))))))..)))))))...))))))))))) ( -56.00) >DroYak_CAF1 63679 104 - 1 GGCGGCUGCCAAUGGAGGGAGCAGGGCAGGAUGCCCACCAAUCCCAGUCCCGCGGCGGGACUGGUCGGUACUCCGGUCGACAGGUUCGCUAUUUUUGGCUCCUC ...((..(((((...((.((((.((((.....))))(((....(((((((((...)))))))))..((....)))))......)))).))....))))).)).. ( -45.50) >consensus GGCGGCUGCCAAUGGAGCGAGCAGGGCAGGAUGCCCACGAUUCCCAGUCCCGAGGCGGGACUGGUCGAUACUCCGGUCGACAGGUUCGCUAUUUUGGCCGCGAC ....((.(((((...(((((((.((((.....))))........(((((((.....)))))))((((((......))))))..)))))))...))))).))... (-40.52 = -41.97 + 1.45)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:22:09 2006