
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 23,837,010 – 23,837,221 |
| Length | 211 |
| Max. P | 0.770489 |

| Location | 23,837,010 – 23,837,100 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 75.64 |
| Mean single sequence MFE | -33.20 |
| Consensus MFE | -18.02 |
| Energy contribution | -17.94 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.54 |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.690545 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 23837010 90 + 27905053 AGCUGUUGCAUC-UG----UA------UGUGUGUGUGUGUGUGUGGAUGCAGCAAA--------UUCCGG----CGCACAGGUGACCGACACCUGCCAGCAGUUACAUAAGAU .(((((((((((-..----((------((..(....)..))))..))))))))...--------....((----(....(((((.....)))))))))))............. ( -31.40) >DroPse_CAF1 47021 99 + 1 AGCUGCUGCCUGCCG----U----------GUGUGUGUGUGGCAGCAUCCAGCUAUCGGCCAUGUGGCGGCUGUGGCACAGGUGAGCUGCAGCUGUCAGCCGUAACAUAAGAU (((((.(((.(((((----.----------.(....)..))))))))..)))))..((((....(((((((((..((........))..)))))))))))))........... ( -45.00) >DroSec_CAF1 42683 86 + 1 AGCUGUUGCAUC-UG----U----------GUGCGUGUGUGUGUGGAUGCAGCAAA--------UUCCGG----CGCACAGGUGACCGGCACCUGCCAGCAGUUACAUAAGAU .((((.((((((-..----(----------(..(....)..))..))))))(((..--------..((((----(((....))).))))....)))))))............. ( -31.00) >DroSim_CAF1 42151 82 + 1 AGCUGUUGCAUC-UG----U----------GU----GUGUGUAUGGAUGCAGCAAA--------UUCCGG----CGCACAGGUGACCGGCACCUGCCAGCAGUUACAUAAGAU (((((((.((((-((----(----------((----((.....((((.........--------.)))))----))))))))))...(((....))))))))))......... ( -29.30) >DroEre_CAF1 46338 100 + 1 AGCUGUUGCAUU-GCCAUGUCAGCAUCUGUGUGUGUGUGUGUGUGGAUGCAGCAAA--------UUCCGG----CGCACAGGUGACCGGCACCUGCCAGCAGUUACAUAAGAU ((((((((((.(-(((.(((..(((((..((..(....)..))..))))).)))..--------....((----(((....))).))))))..)).))))))))......... ( -32.80) >DroYak_CAF1 43357 94 + 1 AGCUGUUGCAUU-GCUAAGUAAGCAUCUCUGUG------UGUGUGGAUGCAGCAAA--------UUCCGG----CGCACAGGUGACCGGCACCUGCCAGCAGUUACAUAAGAU .(((((..(..(-(((.....))))...(((((------(((.((((.........--------.)))))----))))))))..)).(((....))))))............. ( -29.70) >consensus AGCUGUUGCAUC_UG____U__________GUGUGUGUGUGUGUGGAUGCAGCAAA________UUCCGG____CGCACAGGUGACCGGCACCUGCCAGCAGUUACAUAAGAU .(((((((((((.................................)))))))).........................((((((.....))))))..)))............. (-18.02 = -17.94 + -0.08)

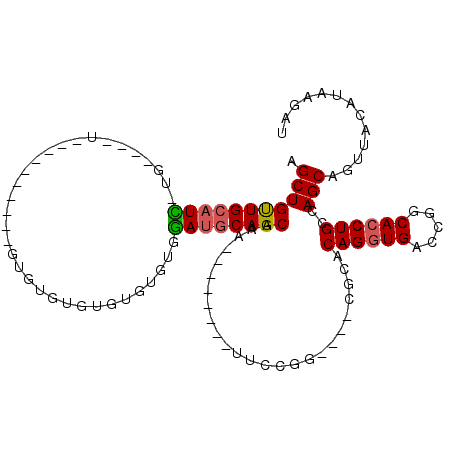
| Location | 23,837,010 – 23,837,100 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 75.64 |
| Mean single sequence MFE | -27.67 |
| Consensus MFE | -17.00 |
| Energy contribution | -17.53 |
| Covariance contribution | 0.53 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.770489 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 23837010 90 - 27905053 AUCUUAUGUAACUGCUGGCAGGUGUCGGUCACCUGUGCG----CCGGAA--------UUUGCUGCAUCCACACACACACACACACA------UA----CA-GAUGCAACAGCU .............((((((((((..((((((....)).)----)))..)--------)))))((((((..................------..----..-)))))).)))). ( -24.15) >DroPse_CAF1 47021 99 - 1 AUCUUAUGUUACGGCUGACAGCUGCAGCUCACCUGUGCCACAGCCGCCACAUGGCCGAUAGCUGGAUGCUGCCACACACACAC----------A----CGGCAGGCAGCAGCU (((((((((..((((((...((.((((.....))))))..))))))..))))))..)))(((((..(((((((..........----------.----.)))).))).))))) ( -34.40) >DroSec_CAF1 42683 86 - 1 AUCUUAUGUAACUGCUGGCAGGUGCCGGUCACCUGUGCG----CCGGAA--------UUUGCUGCAUCCACACACACACGCAC----------A----CA-GAUGCAACAGCU .............((((((((((.(((((((....)).)----)))).)--------)))))((((((...............----------.----..-)))))).)))). ( -26.67) >DroSim_CAF1 42151 82 - 1 AUCUUAUGUAACUGCUGGCAGGUGCCGGUCACCUGUGCG----CCGGAA--------UUUGCUGCAUCCAUACACAC----AC----------A----CA-GAUGCAACAGCU .............((((((((((.(((((((....)).)----)))).)--------)))))((((((.........----..----------.----..-)))))).)))). ( -26.94) >DroEre_CAF1 46338 100 - 1 AUCUUAUGUAACUGCUGGCAGGUGCCGGUCACCUGUGCG----CCGGAA--------UUUGCUGCAUCCACACACACACACACACAGAUGCUGACAUGGC-AAUGCAACAGCU ......((((..(((((((((((.(((((((....)).)----)))).)--------))))))(((((..................)))))......)))-).))))...... ( -26.57) >DroYak_CAF1 43357 94 - 1 AUCUUAUGUAACUGCUGGCAGGUGCCGGUCACCUGUGCG----CCGGAA--------UUUGCUGCAUCCACACA------CACAGAGAUGCUUACUUAGC-AAUGCAACAGCU ......((((..(((((((((((.(((((((....)).)----)))).)--------))))))((((((.....------....).)))))......)))-).))))...... ( -27.30) >consensus AUCUUAUGUAACUGCUGGCAGGUGCCGGUCACCUGUGCG____CCGGAA________UUUGCUGCAUCCACACACACACACAC__________A____CA_GAUGCAACAGCU .............(((((((((((.....)))))))..........................((((((.................................)))))).)))). (-17.00 = -17.53 + 0.53)



| Location | 23,837,100 – 23,837,190 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 90 |
| Reading direction | forward |
| Mean pairwise identity | 92.22 |
| Mean single sequence MFE | -25.50 |
| Consensus MFE | -20.75 |
| Energy contribution | -22.25 |
| Covariance contribution | 1.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.561436 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 23837100 90 + 27905053 UCGCUUAAUUAAACAGACAAACCGUGGGCCAAGAGUGCCAGCUGCAUUAACAGCCCGCGGCGGAAGCAAAUCCAUCCAGAACAGGCAUCU ..((((...............((((((((....(((((.....)))))....)))))))).(((......))).........)))).... ( -27.90) >DroSec_CAF1 42769 90 + 1 UCGCUUAAUUAAACAGACAAACCGUGGGCCAAGAGUGCCAGCUGCAUUAACAGCCCGCGGCGGAAGCAAAUCCAUCCAGAACAGCCAUCU ..(((................((((((((....(((((.....)))))....)))))))).(((......))).........)))..... ( -25.80) >DroSim_CAF1 42233 90 + 1 UCGCUUAAUUAAACAGACAAACCGUGGGCCAAGAGUGCCAGCUGCAUUAACAGCCCGCGGCGGAAGCAAAUCCAUCCAGAACAGGCAUCU ..((((...............((((((((....(((((.....)))))....)))))))).(((......))).........)))).... ( -27.90) >DroEre_CAF1 46438 90 + 1 UCGCUUAAUUAAACAGACAAACCGUGGGCCAAGAGUGCCAGCUGCAUUAACAGCCCGCGGCGGAAGCAAAUCCAUUCAGAACAGGCAUCU ..((((...............((((((((....(((((.....)))))....)))))))).(((......))).........)))).... ( -27.90) >DroYak_CAF1 43451 90 + 1 UCGCUUAAUUAAACAGACAAACCGAGGGCCAAGAGUGCCAGCUGCAUUAACAGCCCGCGGCGGAAGCAAAUCCAUUCAGCACAGGCAUCU ..((((...............(((.((((....(((((.....)))))....)))).))).(((......))).........)))).... ( -24.10) >DroAna_CAF1 43498 90 + 1 UCGCAUAAUUAAACAGACAAACCAUGGGCCAAGCGCAGCAGCUGCAUUAACAGCCCGCGGCGGAAGCAAAAUAACGCACAUCAGUCCGAU ........................(((((...(.((.((.((((......))))..)).((....))........)).)....))))).. ( -19.40) >consensus UCGCUUAAUUAAACAGACAAACCGUGGGCCAAGAGUGCCAGCUGCAUUAACAGCCCGCGGCGGAAGCAAAUCCAUCCAGAACAGGCAUCU ..((((................(((((((....(((((.....)))))....)))))))((....))...............)))).... (-20.75 = -22.25 + 1.50)



| Location | 23,837,130 – 23,837,221 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 82.38 |
| Mean single sequence MFE | -30.60 |
| Consensus MFE | -21.11 |
| Energy contribution | -22.12 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.574720 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 23837130 91 + 27905053 AAGAGUGCCAGCUGCAUUAACAGCCCGCGGCGGAAGCAAAUCCAUCCAGAACAGGCAUCUCUAAAAGUAGGAG---------UAGCCCACGUUGGGGCGC .((((((((.(((((...........)))))(((......)))..........)))).))))...........---------..((((......)))).. ( -29.20) >DroSec_CAF1 42799 91 + 1 AAGAGUGCCAGCUGCAUUAACAGCCCGCGGCGGAAGCAAAUCCAUCCAGAACAGCCAUCUCUAAAAGUAGGAG---------UUGCCUCCUUUCGGGCGC ....(((((.(((((...........)))))(((.((((.(((..........................))).---------)))).))).....))))) ( -25.47) >DroSim_CAF1 42263 91 + 1 AAGAGUGCCAGCUGCAUUAACAGCCCGCGGCGGAAGCAAAUCCAUCCAGAACAGGCAUCUCUAAAAGUAGGAG---------UCGCCUCCUUUGGGGCGC ....(((((.(((((...........)))))(((......)))..(((((..((((..((((.......))))---------..))))..)))))))))) ( -33.30) >DroEre_CAF1 46468 100 + 1 AAGAGUGCCAGCUGCAUUAACAGCCCGCGGCGGAAGCAAAUCCAUUCAGAACAGGCAUCUCUAAAAGUAGGAGUCGAAAGCGUUGCCUACUUUGGGGCGC .((((((((.(((((...........)))))(((......)))..........)))).)))).((((((((.(.(....))....))))))))....... ( -32.70) >DroYak_CAF1 43481 100 + 1 AAGAGUGCCAGCUGCAUUAACAGCCCGCGGCGGAAGCAAAUCCAUUCAGCACAGGCAUCUCUAAAAGUAGGAGACGAAAGCGUUGCCUACUUUGGGGCGC .((((((((.(((((...........)))))(((......)))..........)))).)))).((((((((..(((....)))..))))))))....... ( -35.20) >DroAna_CAF1 43528 94 + 1 AAGCGCAGCAGCUGCAUUAACAGCCCGCGGCGGAAGCAAAAUAACGCACAUCAGUCCGAUGUCAGAGU------CAAAAGUGUAGCCUACUUUGCAGCGC ..((((.(((((((((((........(((((....)).......)))(((((.....)))))......------....))))))))......))).)))) ( -27.71) >consensus AAGAGUGCCAGCUGCAUUAACAGCCCGCGGCGGAAGCAAAUCCAUCCAGAACAGGCAUCUCUAAAAGUAGGAG_________UUGCCUACUUUGGGGCGC .((((((((.((((......)))).....((....))................)))).))))......................((((......)))).. (-21.11 = -22.12 + 1.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:22:02 2006