
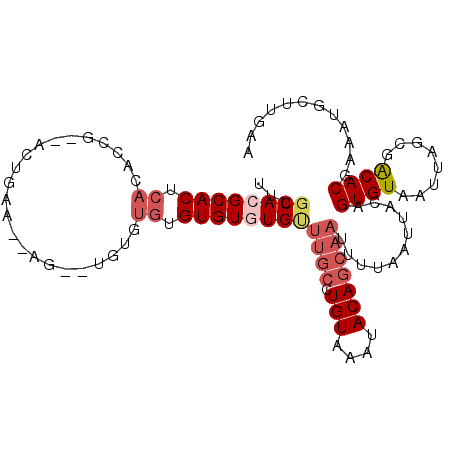
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 23,828,262 – 23,828,362 |
| Length | 100 |
| Max. P | 0.627136 |

| Location | 23,828,262 – 23,828,362 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 80.52 |
| Mean single sequence MFE | -28.37 |
| Consensus MFE | -14.44 |
| Energy contribution | -15.55 |
| Covariance contribution | 1.11 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.51 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.627136 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 23828262 100 + 27905053 UUUGCACGCACUCACACCG--ACUGAACGAG----UGUGUGUGUGUGUUUGCCUGUAAAUACAGCAAUUUUAAUUACAGUGUAAUUAGCGACACGAAAUGCUUGCC ...((((((((.(((((((--......)).)----)))).))))))))((((.(((....)))))))...(((((((...)))))))((((((.....)).)))). ( -30.10) >DroPse_CAF1 37589 94 + 1 UUUGCACGCACUCAGAGUG----------UG--UGCAUGUGUGUAUGCUUGCCUGUAAAUACAGCAAUUUUAAUUACAGUGUAAUUAGCGGCACGAACGGCACGAA ..(((((((((.....)))----------))--))))..(((((...(.(((((((....)))......((((((((...)))))))).)))).)....))))).. ( -28.90) >DroSec_CAF1 34026 104 + 1 UUUGCACGCACUCACACCGAUACUGAACAAG--UGUGUGUGUGUGUGUUUGCCUGUAAAUACAGCAAUUUUAAUUACAGUGUAAUUAGCGACACGAAAUGCUUGCC .((((((((((.(((((..(((((.....))--)))..))))).))))((((.(((....)))))))...........))))))...((((((.....)).)))). ( -28.70) >DroSim_CAF1 33408 106 + 1 UUUGCACGCACUCACACCGAGACUGAACGCGCAUGUGUGUGUGUGUGUUUGCCUGUAAAUACAGCAAUUUUAAUUACAGUGUAAUUAGCGACACGAAAUGCUUGCC ...((((((((.(((((((.(......).))...))))).))))))))((((.(((....)))))))...(((((((...)))))))((((((.....)).)))). ( -27.50) >DroYak_CAF1 34508 88 + 1 UUUGCACGCACUCACACCGAAACUGUG------------UGUGUGUG------UGUAAAUACAGCAAUUUUAAUUACAGUGUAAUUAGCGACACGAAAUGCUUAAA (((((((((((.(((((.......)))------------)).)))))------))))))...((((.((((((((((...))))))).......))).)))).... ( -26.11) >DroPer_CAF1 37755 94 + 1 UUUGCACGCACUCAGAGUG----------UG--UGCAUGUGUGUAUGCUUGCCUGUAAAUACAGCAAUUUUAAUUACAGUGUAAUUAGCGGCACGAACGGCACGAA ..(((((((((.....)))----------))--))))..(((((...(.(((((((....)))......((((((((...)))))))).)))).)....))))).. ( -28.90) >consensus UUUGCACGCACUCACACCG__ACUGAA__AG__UGUGUGUGUGUGUGUUUGCCUGUAAAUACAGCAAUUUUAAUUACAGUGUAAUUAGCGACACGAAAUGCUUGAA ...((((((((.((.......................)).))))))))((((.(((....)))))))...........((((........))))............ (-14.44 = -15.55 + 1.11)


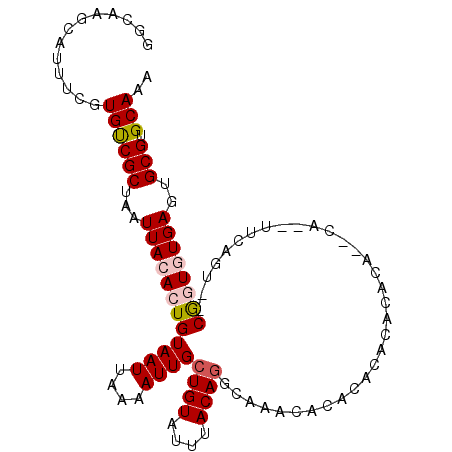
| Location | 23,828,262 – 23,828,362 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 80.52 |
| Mean single sequence MFE | -25.50 |
| Consensus MFE | -13.64 |
| Energy contribution | -14.03 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.54 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 23828262 100 - 27905053 GGCAAGCAUUUCGUGUCGCUAAUUACACUGUAAUUAAAAUUGCUGUAUUUACAGGCAAACACACACACACA----CUCGUUCAGU--CGGUGUGAGUGCGUGCAAA .(((.(((((..((((...(((((((...)))))))...(((((((....))).)))))))).....((((----(.((......--)))))))))))).)))... ( -27.90) >DroPse_CAF1 37589 94 - 1 UUCGUGCCGUUCGUGCCGCUAAUUACACUGUAAUUAAAAUUGCUGUAUUUACAGGCAAGCAUACACACAUGCA--CA----------CACUCUGAGUGCGUGCAAA ...(((..((...((((..(((.((((..(((((....))))))))).)))..)))).....)).))).((((--(.----------(((.....))).))))).. ( -25.70) >DroSec_CAF1 34026 104 - 1 GGCAAGCAUUUCGUGUCGCUAAUUACACUGUAAUUAAAAUUGCUGUAUUUACAGGCAAACACACACACACACA--CUUGUUCAGUAUCGGUGUGAGUGCGUGCAAA .(((.(((((..((((...(((((((...)))))))...(((((((....))).)))))))).......((((--((.((.....)).))))))))))).)))... ( -26.90) >DroSim_CAF1 33408 106 - 1 GGCAAGCAUUUCGUGUCGCUAAUUACACUGUAAUUAAAAUUGCUGUAUUUACAGGCAAACACACACACACACAUGCGCGUUCAGUCUCGGUGUGAGUGCGUGCAAA .(((.(((((..((((...(((((((...)))))))...(((((((....))).)))))))).....(((((....((.....))....)))))))))).)))... ( -27.10) >DroYak_CAF1 34508 88 - 1 UUUAAGCAUUUCGUGUCGCUAAUUACACUGUAAUUAAAAUUGCUGUAUUUACA------CACACACA------------CACAGUUUCGGUGUGAGUGCGUGCAAA .....((((...((((........)))).(((((....))))).(((((((((------(....((.------------....))....))))))))))))))... ( -19.70) >DroPer_CAF1 37755 94 - 1 UUCGUGCCGUUCGUGCCGCUAAUUACACUGUAAUUAAAAUUGCUGUAUUUACAGGCAAGCAUACACACAUGCA--CA----------CACUCUGAGUGCGUGCAAA ...(((..((...((((..(((.((((..(((((....))))))))).)))..)))).....)).))).((((--(.----------(((.....))).))))).. ( -25.70) >consensus GGCAAGCAUUUCGUGUCGCUAAUUACACUGUAAUUAAAAUUGCUGUAUUUACAGGCAAACACACACACACACA__CA__UUCAGU__CGGUGUGAGUGCGUGCAAA .............((((((...((((((((((((....))))((((....)))).................................))))))))..))).))).. (-13.64 = -14.03 + 0.39)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:21:54 2006