
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
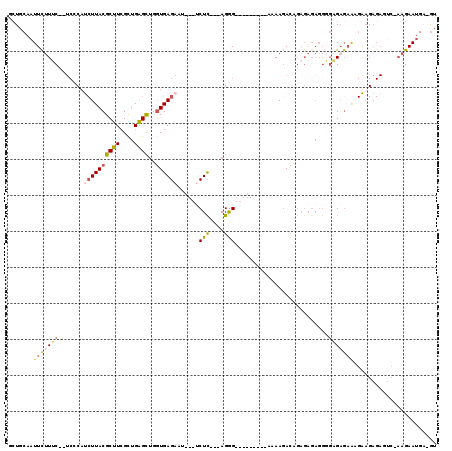
| Location | 23,814,912 – 23,815,111 |
| Length | 199 |
| Max. P | 0.944653 |

| Location | 23,814,912 – 23,815,016 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 66.82 |
| Mean single sequence MFE | -29.10 |
| Consensus MFE | -6.28 |
| Energy contribution | -7.36 |
| Covariance contribution | 1.08 |
| Combinations/Pair | 1.35 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.22 |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.815088 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
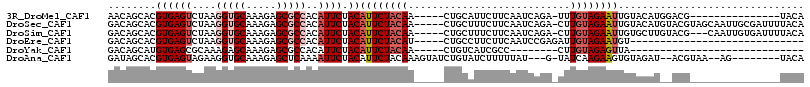
>3R_DroMel_CAF1 23814912 104 + 27905053 AAUGCAAUUCUUUCUCUCCCAUCUUACGCUUCGCUGAGCUGGUGAGAAU---UCUU---AGGG---------AAAAGAUAGAGAGAGGGGAGAGAAAGAAGAGAGUCGAAGAAUGAGGU .......(((((((((((((.((((.(.((((.(((((..(((....))---))))---)).)---------)..))...).)))).)))))))))))))................... ( -32.20) >DroSec_CAF1 20123 81 + 1 GCUGCAAUUCUUUC---------UUACGCUUCGCUGAGCUGGUGAGAAU---UCUC---AGGG---------AAAAGACAGAGAGAGGGGAGAGAA-------------AGAAUGU-GU ...(((((((((((---------((.(.((((.(((..((..((((...---.)))---)...---------...)).)))...)))).).)))))-------------))))).)-)) ( -23.90) >DroSim_CAF1 20178 101 + 1 GCUGCAAUUCUUUC--UCCCAUCUUACGCUUCGCUGAGCUGGUGAGAAU---UCUC---AGGG---------AAAAGACAGAGAGAGGGGAGAGAAGGAAGAGAGUCCAAGAAUGA-GU (((.((.(((((((--(((..((((...((((.(((((..(((....))---))))---)).)---------)..))...))))..))))))))))(((......))).....)))-)) ( -29.50) >DroYak_CAF1 20669 91 + 1 ACUGCAAUUUCUGC--UCUCAUCUUACGCUUCGCUGAGCUGCUGAGAAA---ACUC---AGAG---------AAAAGAGA-----------GAGAGAGAAGAGAGUCGAAGAAUGAGGU (((.((.((((.((--((((.((((.(..(((.((...((.(((((...---.)))---))))---------...)).))-----------).).)))).)))))).))))..)).))) ( -27.00) >DroAna_CAF1 20894 111 + 1 GCUGCACUUUUUGC--UCUCACCUUACGCUUGGUUGGGUGUGUGAGAAAAACGCUCGAGAGGGCACAUUAAUGAAAAACAGAGAGAGGUGCGC--UCGGAGAGAG---AAGAACGG-AA ...(((((((((..--((((((...((((((....)))))))))))).....((((....))))..................))))))))).(--((.....)))---........-.. ( -32.90) >consensus GCUGCAAUUCUUUC__UCCCAUCUUACGCUUCGCUGAGCUGGUGAGAAU___UCUC___AGGG_________AAAAGACAGAGAGAGGGGAGAGAAAGAAGAGAGUC_AAGAAUGA_GU .......(((((((.......((((((((((....))))..))))))......(((....)))..........................)))))))....................... ( -6.28 = -7.36 + 1.08)


| Location | 23,814,912 – 23,815,016 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 66.82 |
| Mean single sequence MFE | -22.60 |
| Consensus MFE | -5.26 |
| Energy contribution | -6.40 |
| Covariance contribution | 1.14 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.23 |
| SVM decision value | 0.80 |
| SVM RNA-class probability | 0.854412 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 23814912 104 - 27905053 ACCUCAUUCUUCGACUCUCUUCUUUCUCUCCCCUCUCUCUAUCUUUU---------CCCU---AAGA---AUUCUCACCAGCUCAGCGAAGCGUAAGAUGGGAGAGAAAGAAUUGCAUU ...................(((((((((((((.(((..(..((((..---------....---))))---..........(((......))))..))).)))))))))))))....... ( -28.60) >DroSec_CAF1 20123 81 - 1 AC-ACAUUCU-------------UUCUCUCCCCUCUCUCUGUCUUUU---------CCCU---GAGA---AUUCUCACCAGCUCAGCGAAGCGUAA---------GAAAGAAUUGCAGC .(-(.(((((-------------((((..(..((.((.(((......---------...(---(((.---...))))......))).)))).)..)---------)))))))))).... ( -14.16) >DroSim_CAF1 20178 101 - 1 AC-UCAUUCUUGGACUCUCUUCCUUCUCUCCCCUCUCUCUGUCUUUU---------CCCU---GAGA---AUUCUCACCAGCUCAGCGAAGCGUAAGAUGGGA--GAAAGAAUUGCAGC ..-..(((((((((......)))...((((((.(((..(.((...((---------(.((---(((.---...........))))).))))))..))).))))--))))))))...... ( -24.50) >DroYak_CAF1 20669 91 - 1 ACCUCAUUCUUCGACUCUCUUCUCUCUC-----------UCUCUUUU---------CUCU---GAGU---UUUCUCAGCAGCUCAGCGAAGCGUAAGAUGAGA--GCAGAAAUUGCAGU ........((.(((...(((.(((((((-----------(..(.(((---------(.((---((((---(........))))))).)))).)..))).))))--).)))..))).)). ( -24.30) >DroAna_CAF1 20894 111 - 1 UU-CCGUUCUU---CUCUCUCCGA--GCGCACCUCUCUCUGUUUUUCAUUAAUGUGCCCUCUCGAGCGUUUUUCUCACACACCCAACCAAGCGUAAGGUGAGA--GCAAAAAGUGCAGC ..-..((.(((---(((((.((((--(.((((.......((.....)).....)))).)))....((((((.................))))))..)).))))--)....))).))... ( -21.43) >consensus AC_UCAUUCUU_GACUCUCUUCUUUCUCUCCCCUCUCUCUGUCUUUU_________CCCU___GAGA___AUUCUCACCAGCUCAGCGAAGCGUAAGAUGAGA__GAAAGAAUUGCAGC .....((((((....((((.((.........................................(((.......)))....(((......)))....)).))))....))))))...... ( -5.26 = -6.40 + 1.14)

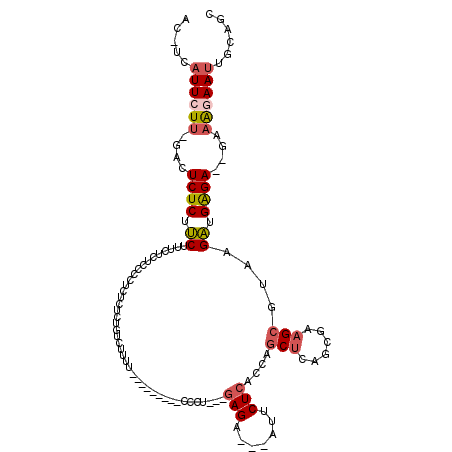

| Location | 23,814,977 – 23,815,086 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 73.01 |
| Mean single sequence MFE | -29.20 |
| Consensus MFE | -10.01 |
| Energy contribution | -10.20 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.34 |
| SVM decision value | 0.77 |
| SVM RNA-class probability | 0.847444 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 23814977 109 + 27905053 GAGAGAGGGGAGAGAAAGAAGAGAGUCGAAGAAUGAGGUAACAGCACGUGAGUCUAAGGUGCAAAGAGCGCCACAUUCUACAUUCUACAA-----CUGCAUUCUUCAAUCAGA-U ....((..((((((..((..((((((.(.((((((.......((.((....))))..(((((.....))))).)))))).)))))).)..-----))...))))))..))...-. ( -30.70) >DroSec_CAF1 20179 95 + 1 GAGAGAGGGGAGAGAA-------------AGAAUGU-GUGACAGCACGUGAGUCUAAGGUGCAAAGAGCGCCACAUUCUACAUUCUACAA-----CUGCUUUCUUCAAUCAGA-C ((..((((..((((..-------------(((((((-(.((.((.((....))))..(((((.....)))))....))))))))))....-----)).))..))))..))...-. ( -30.50) >DroSim_CAF1 20241 108 + 1 GAGAGAGGGGAGAGAAGGAAGAGAGUCCAAGAAUGA-GUGACAGCACGUGAGUCUAAGGUGCAAAGAGCGCCACAUUCUACAUUCUACAA-----CUGCUUUCUUCAAUCAGA-C ..((..((((((((.((....(((((...((((((.-.....((.((....))))..(((((.....))))).))))))..)))))....-----)).))))))))..))...-. ( -34.00) >DroEre_CAF1 20691 103 + 1 CAGA-------GAGAAAGAAGAGAGUCGAAGAAUGAGGUGACAGCACGUGAGUCUAAGGUGCAAAGAGCGCCACAUUCUACAUUCUACAU-----CUGCCUUCUUCAAUCCGAGA ..((-------..(((.((((..((..(.(((((((((((..((.((....))))..(((((.....))))).)).))).)))))).)..-----))..)))))))..))..... ( -28.30) >DroYak_CAF1 20732 91 + 1 -----------GAGAGAGAAGAGAGUCGAAGAAUGAGGUGACAGCAUGUGAGCGCAAAGAGCAAAGAGCGCCACAUUCUACAUUCUACAA-----CUGUCAUCGCC--------C -----------......((.((.(((.(.((((((.(....)((.(((((.((((............))))))))).)).)))))).).)-----)).)).))...--------. ( -27.90) >DroAna_CAF1 20972 105 + 1 GAGAGAGGUGCGC--UCGGAGAGAG---AAGAACGG-AAGAUAGCACGUGAGUAGAAGGUGCAAAGAGCUCAAAAUUCUACAUUCUACAAAGUAUCUGUAUCUUUUUAU---G-U .((((((((((.(--((.....)))---.(((((..-..)...........((((((.(.((.....)).)....)))))).))))...........))))))))))..---.-. ( -23.80) >consensus GAGAGAGGGGAGAGAAAGAAGAGAGUCGAAGAAUGA_GUGACAGCACGUGAGUCUAAGGUGCAAAGAGCGCCACAUUCUACAUUCUACAA_____CUGCAUUCUUCAAUCAGA_C .............................((((((........((((.(.......).))))..((((.......)))).))))))............................. (-10.01 = -10.20 + 0.19)


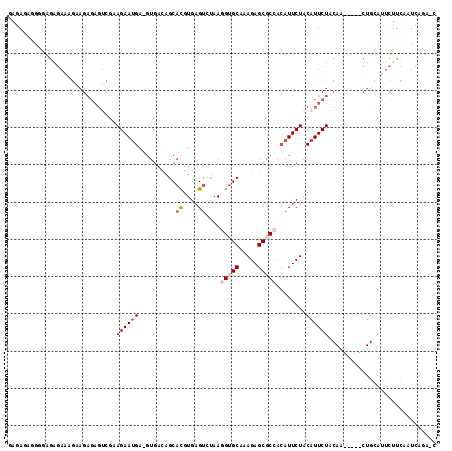
| Location | 23,815,016 – 23,815,111 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 68.00 |
| Mean single sequence MFE | -24.52 |
| Consensus MFE | -9.27 |
| Energy contribution | -10.46 |
| Covariance contribution | 1.19 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.38 |
| SVM decision value | 1.35 |
| SVM RNA-class probability | 0.944653 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 23815016 95 + 27905053 AACAGCACGUGAGUCUAAGGUGCAAAGAGCGCCACAUUCUACAUUCUACAA-----CUGCAUUCUUCAAUCAGA-UUUGUAGAAUUGUACAUGGACG---------------UACA ........(((.(((((.(((((.....)))))......((((((((((((-----(((.(((....)))))).-.)))))))).))))..))))).---------------))). ( -27.10) >DroSec_CAF1 20204 110 + 1 GACAGCACGUGAGUCUAAGGUGCAAAGAGCGCCACAUUCUACAUUCUACAA-----CUGCUUUCUUCAAUCAGA-CUUGUAGAAUUGUACAUGUACGUAGCAAUUGCGAUUUUACA ....((((((........(((((.....)))))((((..((((((((((((-----(((.((.....)).))).-.)))))))).)))).)))))))).))............... ( -27.10) >DroSim_CAF1 20279 107 + 1 GACAGCACGUGAGUCUAAGGUGCAAAGAGCGCCACAUUCUACAUUCUACAA-----CUGCUUUCUUCAAUCAGA-CUUGUAGAAUUGUGCUUGUACG---CAAUUGUGAUUUUACA .(((((((((((((....(((((.....)))))..)))).))(((((((((-----(((.((.....)).))).-.))))))))).)))).))).((---(....)))........ ( -27.50) >DroEre_CAF1 20723 82 + 1 GACAGCACGUGAGUCUAAGGUGCAAAGAGCGCCACAUUCUACAUUCUACAU-----CUGCCUUCUUCAAUCCGAGAUUGUAGAAUGU----------------------------- ...((.((....))))..(((((.....)))))((((((((((.(((....-----.((.......)).....))).))))))))))----------------------------- ( -24.80) >DroYak_CAF1 20760 74 + 1 GACAGCAUGUGAGCGCAAAGAGCAAAGAGCGCCACAUUCUACAUUCUACAA-----CUGUCAUCGCC--------CUUGUAGAGUUA----------------------------- ...((.(((((.((((............))))))))).))..(((((((((-----..((....)).--------.)))))))))..----------------------------- ( -20.60) >DroAna_CAF1 21005 100 + 1 GAUAGCACGUGAGUAGAAGGUGCAAAGAGCUCAAAAUUCUACAUUCUACAAAGUAUCUGUAUCUUUUUAU---G-UAUCAAGAAGUGUAGAU--ACGUAA--AG--------UACA ........(((.((((((.(.((.....)).)....))))))..........(((((((((.((((((..---.-....)))))))))))))--))....--..--------))). ( -20.00) >consensus GACAGCACGUGAGUCUAAGGUGCAAAGAGCGCCACAUUCUACAUUCUACAA_____CUGCAUUCUUCAAUCAGA_CUUGUAGAAUUGUACAU__ACG_______________UACA ........((((((....(((((.....)))))..)))).))((((((((...........................))))))))............................... ( -9.27 = -10.46 + 1.19)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:21:51 2006