
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 23,808,273 – 23,808,468 |
| Length | 195 |
| Max. P | 0.500000 |

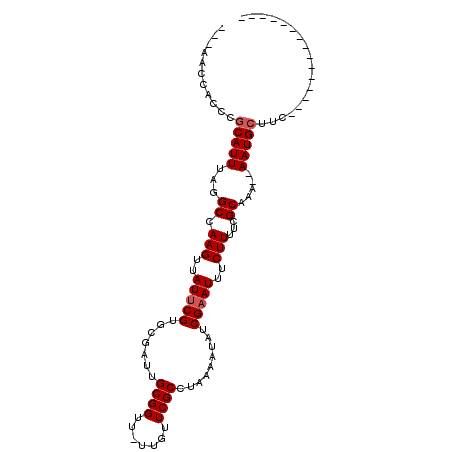
| Location | 23,808,273 – 23,808,373 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 81.04 |
| Mean single sequence MFE | -19.03 |
| Consensus MFE | -13.64 |
| Energy contribution | -13.97 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.14 |
| Structure conservation index | 0.72 |
| SVM decision value | -0.08 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 23808273 100 + 27905053 ---CAUCACCCGCAUUUAGGCCAAGUUAUUCGUUCGAUUGCGGUU-UUGUUCGCCUAAAAUAUCGAAUUUCUUUUGGCAAA--AAUGCUUCUGCUACCCCACCAAC ---........((((((..((((((......(((((((.((((..-....)))).......)))))))....))))))..)--))))).................. ( -23.30) >DroSec_CAF1 13552 85 + 1 ---CAUCGCCCGCAUUUAGGCCAAGUUAUUCGUUCGAUUGCGGUU-UUGUUCGCCUGAAAUAUCGAAUUUCUUUUCGCAAA--AAUGCUUC--------------- ---........((((((..((.(((..(((((((((...((((..-....)))).))))....)))))..)))...))..)--)))))...--------------- ( -18.60) >DroSim_CAF1 13598 85 + 1 ---CAACGCCCGCAUUUAGGCCAAGUUAUGCGUUCGAUUGCGGUU-UUGUUCGCCUGAAAUAUCGAAUUUCUUUUCGCAAA--AAUGCUUC--------------- ---........((((((((((((((...((((......))))..)-)))...)))))))....((((......))))....--..)))...--------------- ( -17.70) >DroEre_CAF1 13804 94 + 1 ---AGCCACCCGCAUUUAGGCCAAGUUAUUCGUGCGAUUGCGGUUUUUGUUCGCCUAAAAUGUCGAAUUUCUUUUUGCAGA--AAUGCUCCUACUCCUC------- ---........((((((..((.(((..(((((.(((.((((((.......))))...)).))))))))..)))...))..)--)))))...........------- ( -19.00) >DroYak_CAF1 14144 89 + 1 ---AGCCACCCGCAUUUCAGCCAAGUUAUUCGUGCGAUUGCGGUU-UUGUUCGCCUAAAAUAUCGAAUUUCUUUUCGCAAA--AAUGCUUCUACU----------- ---........((((((..((.(((..(((((.......((((..-....)))).........)))))..)))...))..)--))))).......----------- ( -17.49) >DroAna_CAF1 14174 90 + 1 AAAAACAAUCCGCAUUUAGGCUAAGUUAUUCGUGCAUUUGCGGUU-UUGUUCGCCUGAAAUAUCGAAUUUCUUUUCGCAAUUUAAUGGUUU--------------- ...(((((.(((((.....((...(.....)..))...)))))..-))))).(((..((((..((((......))))..))))...)))..--------------- ( -18.10) >consensus ___AACCACCCGCAUUUAGGCCAAGUUAUUCGUGCGAUUGCGGUU_UUGUUCGCCUAAAAUAUCGAAUUUCUUUUCGCAAA__AAUGCUUC_______________ ...........(((((...((.(((..(((((.......((((.......)))).........)))))..)))...)).....))))).................. (-13.64 = -13.97 + 0.33)

| Location | 23,808,373 – 23,808,468 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 83.03 |
| Mean single sequence MFE | -15.20 |
| Consensus MFE | -11.89 |
| Energy contribution | -11.95 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.35 |
| Structure conservation index | 0.78 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 23808373 95 + 27905053 CUCACCCCCGCCCCCUCCUGCCACCGGCUGCUGCAAAUAAUAAAACCGAUGUUAAUCCUUCAAGGACAUGCAGCCGGACAUAUGUUUGUCCAAAC .........................((((((((....(((((.......))))).(((.....))))).))))))(((((......))))).... ( -22.40) >DroSec_CAF1 13637 85 + 1 ----------CCCCCUCAUGCCAGCGACUGCCAUAAAUAAUAAAACCGAUGUUAAUCUUUCAAGGACAUGCAGCCGGACAUAUGUUUGUCCAAAC ----------....((((((((.((....))................(((....)))......)).)))).))..(((((......))))).... ( -15.30) >DroSim_CAF1 13683 85 + 1 ----------CCCCCUCAUGCCAACGACUGCCAUAAAUAAUAAAACCGAUGUUAAUCUUUCAAGGACAUGCAGCCGGACAUAUGUUUGUCCAAAC ----------....((((((((...(.....)...............(((....)))......)).)))).))..(((((......))))).... ( -13.30) >DroEre_CAF1 13898 88 + 1 -------CCUACUCCUCCCUCCUCCGCCUGCCAUAAAUAAUAAAACCGAUGUUAAUCUUUCAAGGACAUGCAGCCGGACAUAUGUUUGUCCAAAC -------..................(.((((......(((((.......))))).(((.....)))...)))).)(((((......))))).... ( -12.50) >DroYak_CAF1 14233 79 + 1 ----------------CCCUCCACCGCCUGCCAUAAAUAAUAAAACCGAUGUUAAUCUUUCAAGGACAUGCAGACGGACAUAUGUUUGUCCAAAC ----------------.........(.((((......(((((.......))))).(((.....)))...)))).)(((((......))))).... ( -12.50) >DroAna_CAF1 14264 78 + 1 ----------C------CC-CCCACACCUGCCAUAAAUUAUAAAACCGAUGUUAAUCUUUCAAAGACAGGCAGCGGGACAUAUGUUUGUCCAAAC ----------.------..-......((((((...........(((....)))..(((.....)))..))))).)(((((......))))).... ( -15.20) >consensus __________CCCCCUCCCGCCACCGCCUGCCAUAAAUAAUAAAACCGAUGUUAAUCUUUCAAGGACAUGCAGCCGGACAUAUGUUUGUCCAAAC .........................(.((((......(((((.......))))).(((.....)))...)))).)(((((......))))).... (-11.89 = -11.95 + 0.06)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:21:40 2006